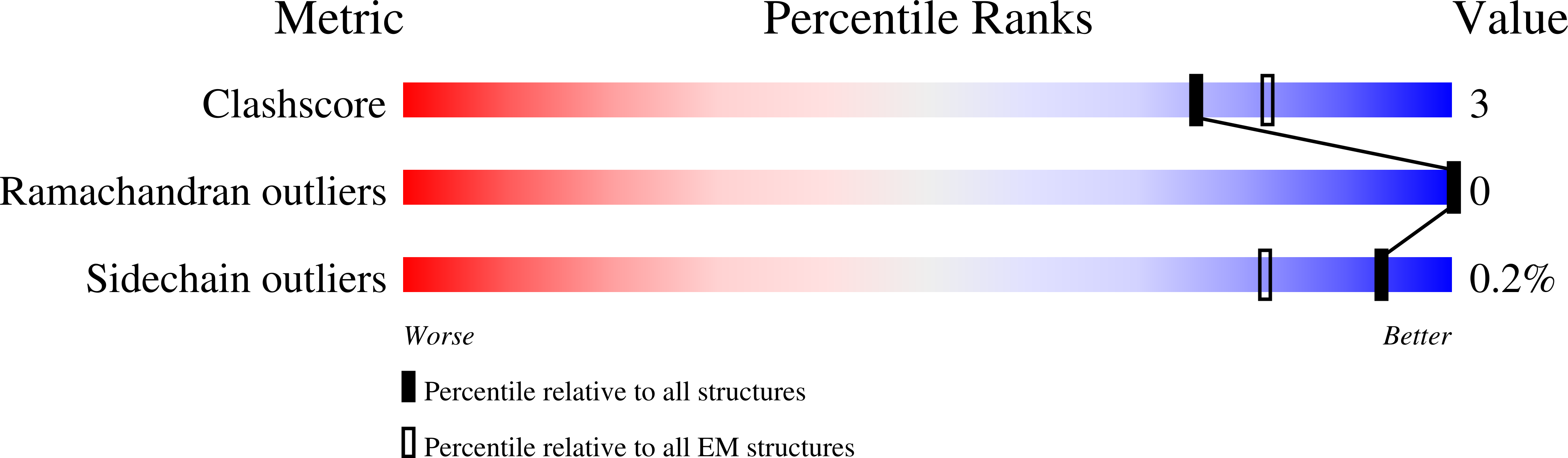SARS-COV-2 Omicron variants conformationally escape a rare quaternary antibody binding mode.
Goike, J., Hsieh, C.L., Horton, A.P., Gardner, E.C., Zhou, L., Bartzoka, F., Wang, N., Javanmardi, K., Herbert, A., Abbassi, S., Xie, X., Xia, H., Shi, P.Y., Renberg, R., Segall-Shapiro, T.H., Terrace, C.I., Wu, W., Shroff, R., Byrom, M., Ellington, A.D., Marcotte, E.M., Musser, J.M., Kuchipudi, S.V., Kapur, V., Georgiou, G., Weaver, S.C., Dye, J.M., Boutz, D.R., McLellan, J.S., Gollihar, J.D.(2023) Commun Biol 6: 1250-1250
- PubMed: 38082099
- DOI: https://doi.org/10.1038/s42003-023-05649-6
- Primary Citation of Related Structures:
8TM1, 8TMA - PubMed Abstract:
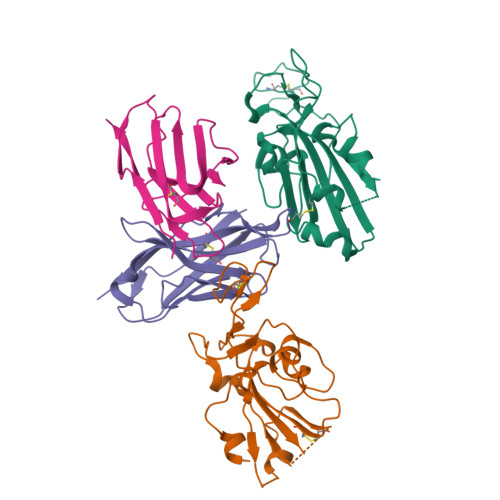
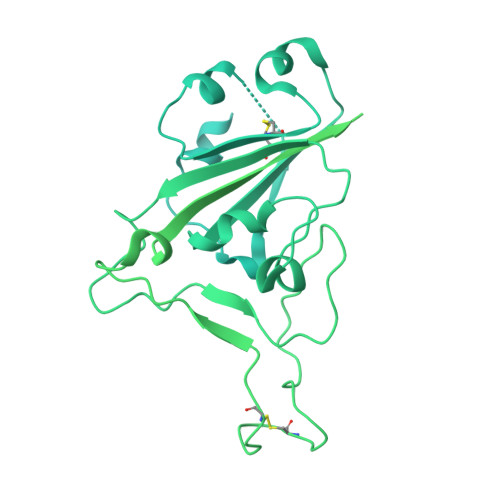
The ongoing evolution of SARS-CoV-2 into more easily transmissible and infectious variants has provided unprecedented insight into mutations enabling immune escape. Understanding how these mutations affect the dynamics of antibody-antigen interactions is crucial to the development of broadly protective antibodies and vaccines. Here we report the characterization of a potent neutralizing antibody (N3-1) identified from a COVID-19 patient during the first disease wave. Cryogenic electron microscopy revealed a quaternary binding mode that enables direct interactions with all three receptor-binding domains of the spike protein trimer, resulting in extraordinary avidity and potent neutralization of all major variants of concern until the emergence of Omicron. Structure-based rational design of N3-1 mutants improved binding to all Omicron variants but only partially restored neutralization of the conformationally distinct Omicron BA.1. This study provides new insights into immune evasion through changes in spike protein dynamics and highlights considerations for future conformationally biased multivalent vaccine designs.
Organizational Affiliation:
Center for Systems and Synthetic Biology, Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX, USA.