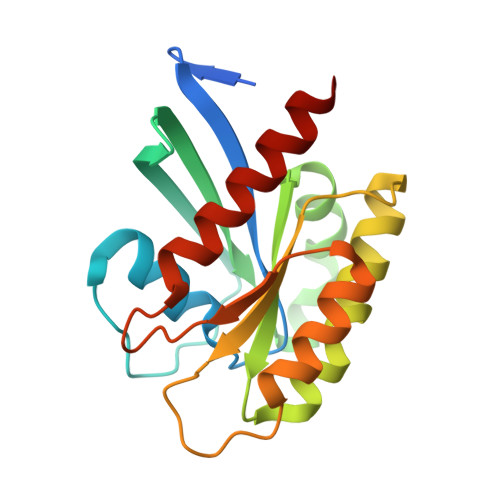
Crystal structure of NRAS Q61K with a ligand-induced pocket near switch II.
Gebregiworgis, T., Chan, J.Y., Kuntz, D.A., Prive, G.G., Marshall, C.B., Ikura, M.(2024) Eur J Cell Biol 103: 151414-151414
- PubMed: 38640594
- DOI: https://doi.org/10.1016/j.ejcb.2024.151414
- Primary Citation of Related Structures:
8VM2 - PubMed Abstract:
The RAS isoforms (KRAS, HRAS and NRAS) have distinct cancer type-specific profiles. NRAS mutations are the second most prevalent RAS mutations in skin and hematological malignancies. Although RAS proteins were considered undruggable for decades, isoform and mutation-specific investigations have produced successful RAS inhibitors that are either specific to certain mutants, isoforms (pan-KRAS) or target all RAS proteins (pan-RAS). While extensive structural and biochemical investigations have focused mainly on K- and H-RAS mutations, NRAS mutations have received less attention, and the most prevalent NRAS mutations in human cancers, Q61K and Q61R, are rare in K- and H-RAS. This manuscript presents a crystal structure of the NRAS Q61K mutant in the GTP-bound form. Our structure reveals a previously unseen pocket near switch II induced by the binding of a ligand to the active form of the protein. This observation reveals a binding site that can potentially be exploited for development of inhibitors against mutant NRAS. Furthermore, the well-resolved catalytic site of this GTPase bound to native GTP provides insight into the stalled GTP hydrolysis observed for NRAS-Q61K.
Organizational Affiliation:
Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario M5G 1L7, Canada; Department of Biochemistry, Schulich School of Medicine and Dentistry, Western University, London, Ontario N6A 5C1, Canada; Department of Oncology, Schulich School of Medicine and Dentistry, Western University, London, Ontario N6A 5W9, Canada. Electronic address: tgebregi@uwo.ca.