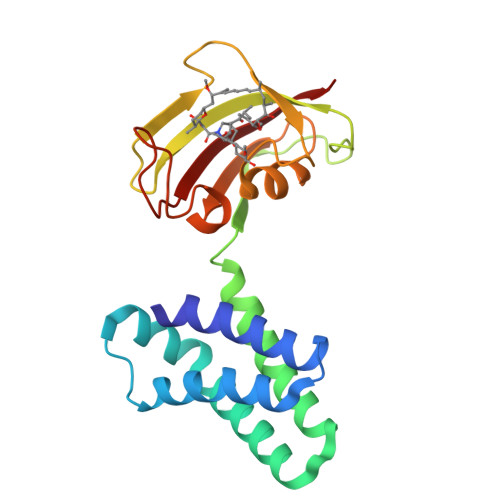
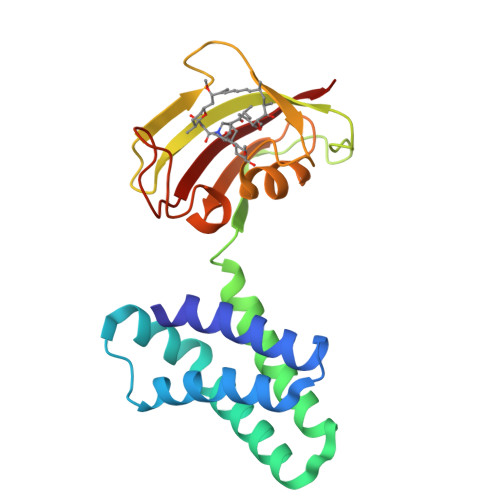
Structural insights into rapamycin-induced oligomerization of a FRB-FKBP fusion protein.
Inobe, T., Sakaguchi, R., Obita, T., Mukaiyama, A., Koike, S., Yokoyama, T., Mizuguchi, M., Akiyama, S.(2024) FEBS Lett 598: 2292-2305
- PubMed: 39031920
- DOI: https://doi.org/10.1002/1873-3468.14986
- Primary Citation of Related Structures:
8XI9 - PubMed Abstract:
Inducible dimerization systems, such as rapamycin-induced dimerization of FK506 binding protein (FKBP) and FKBP-rapamycin binding (FRB) domain, are widely employed chemical biology tools to manipulate cellular functions. We previously advanced an inducible dimerization system into an inducible oligomerization system by developing a bivalent fusion protein, FRB-FKBP, which forms large oligomers upon rapamycin addition and can be used to manipulate cells. However, the oligomeric structure of FRB-FKBP remains unclear. Here, we report that FRB-FKBP forms a rotationally symmetric trimer in crystals, but a larger oligomer in solution, primarily tetramers and pentamers, which maintain similar inter-subunit contacts as in the crystal trimer. These findings expand the applications of the FRB-FKBP oligomerization system in diverse biological events.
Organizational Affiliation:
Graduate School of Science and Engineering, University of Toyama, Japan.