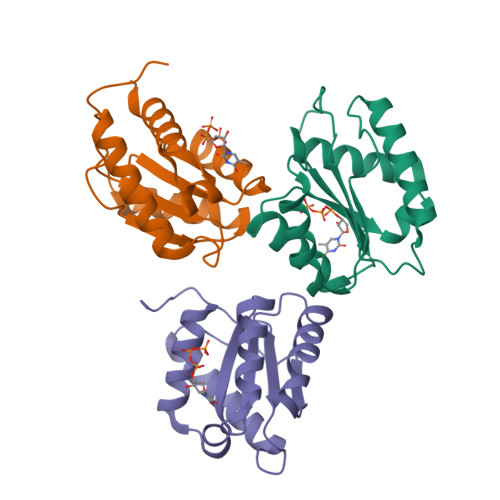
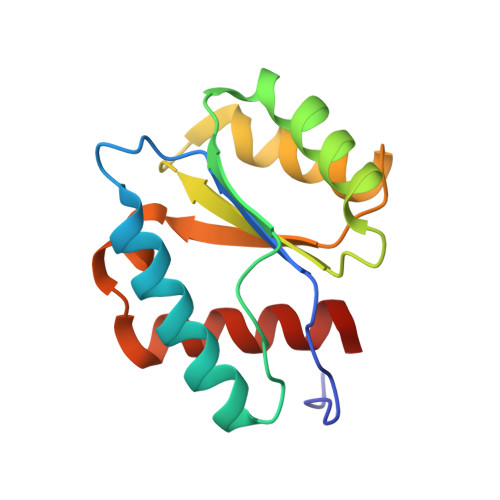
Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Lei, L., Harp, J.M., Chaput, J.C., Wassarman, K., Schlegel, M.K., Manoharan, M., Egli, M.(2024) Curr Protoc 4: e1088-e1088
- PubMed: 38923271
- DOI: https://doi.org/10.1002/cpz1.1088
- Primary Citation of Related Structures:
9BEZ, 9BF0, 9BF2 - PubMed Abstract:
The middle (MID) domain of eukaryotic Argonaute (Ago) proteins and archaeal and bacterial homologues mediates the interaction with the 5'-terminal nucleotide of miRNA and siRNA guide strands. The MID domain of human Ago2 (hAgo2) is comprised of 139 amino acids with a molecular weight of 15.56 kDa. MID adopts a Rossman-like beta1-alpha1-beta2-alpha2-beta3-alpha3-beta4-alpha4 fold with a nucleotide specificity loop between beta3 and alpha3. Multiple crystal structures of nucleotides bound to hAgo2 MID have been reported, whereby complexes were obtained by soaking ligands into crystals of MID domain alone. This protocol describes a simplified one-step approach to grow well-diffracting crystals of hAgo2 MID-nucleotide complexes by mixing purified His 6 -SUMO-MID fusion protein, Ulp1 protease, and excess nucleotide in the presence of buffer and precipitant. The crystal structures of MID complexes with UMP, UTP and 2'-3' linked α-L-threofuranosyl thymidine-3'-triphosphate (tTTP) are presented. This article also describes fluorescence-based assays to measure dissociation constants (K d ) of MID-nucleotide interactions for nucleoside 5'-monophosphates and nucleoside 3',5'-bisphosphates. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC. Basic Protocol 1: Crystallization of Ago2 MID-nucleotide complexes Basic Protocol 2: Measurement of dissociation constant K d between Ago2 MID and nucleotides.
Organizational Affiliation:
Department of Biochemistry, School of Medicine, Vanderbilt University, Nashville, Tennessee.