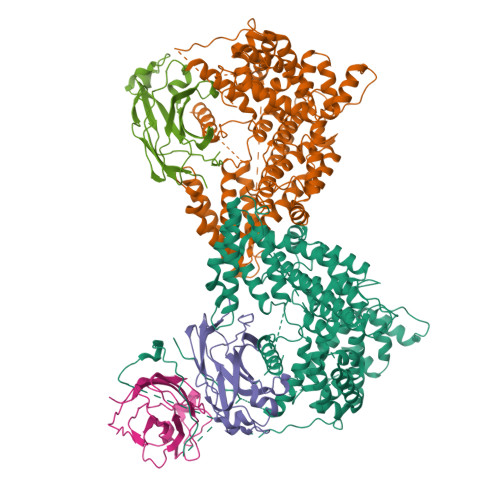
Mechanism of ASF1 engagement by CDAN1.
Sedor, S.F., Shao, S.(2025) Nat Commun 16: 2599-2599
- PubMed: 40091041
- DOI: https://doi.org/10.1038/s41467-025-57950-z
- Primary Citation of Related Structures:
9CVC - PubMed Abstract:
Codanin-1 (CDAN1) is an essential and ubiquitous protein named after congenital dyserythropoietic anemia type I, an autosomal recessive disease that manifests from mutations in CDAN1 or CDIN1 (CDAN1 interacting nuclease 1). CDAN1 interacts with CDIN1 and the paralogous histone H3-H4 chaperones ASF1A (Anti-Silencing Function 1 A) and ASF1B. However, CDAN1 function remains unclear. Here, we analyze CDAN1 complexes using biochemistry, single-particle cryo-EM, and structural predictions. We find that CDAN1 dimerizes and assembles into cytosolic complexes with CDIN1 and multiple copies of ASF1A/B. One CDAN1 can engage two ASF1 through two B-domains commonly found in ASF1 binding partners and two helices that mimic histone H3 binding. We additionally show that ASF1A and ASF1B have different requirements for CDAN1 engagement. Our findings explain how CDAN1 sequesters ASF1A/B by occupying all functional binding sites known to facilitate histone chaperoning and provide molecular-level insights into this enigmatic complex.
Organizational Affiliation:
Department of Cell Biology, Harvard Medical School, Boston, MA, USA.