Crystal structure of synthetic ubiquitin variant R4 designed by ProteinMPNN
Wu, K.-P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ubiquitin variant R4 | 103 | synthetic construct | Mutation(s): 0 | 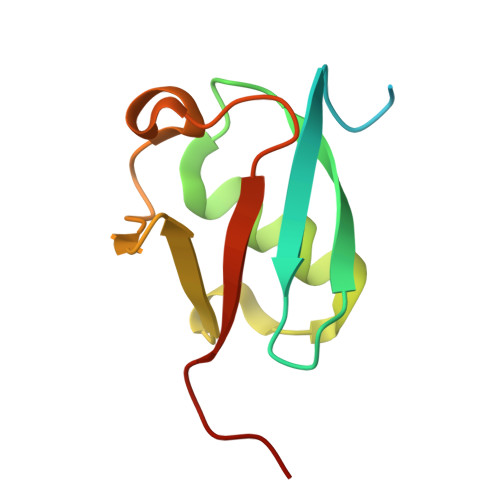 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | C [auth A], D [auth A], E [auth A], F [auth A], G [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 50.845 | α = 90 |
| b = 50.845 | β = 90 |
| c = 102.666 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Coot | model building |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Academia Sinica (Taiwan) | Taiwan | AS-GCS-113-L05 |
| Academia Sinica (Taiwan) | Taiwan | AS-CDA-110-L03 |
| Ministry of Science and Technology (MoST, Taiwan) | Taiwan | NSTC-113-2113-M-001-030 |