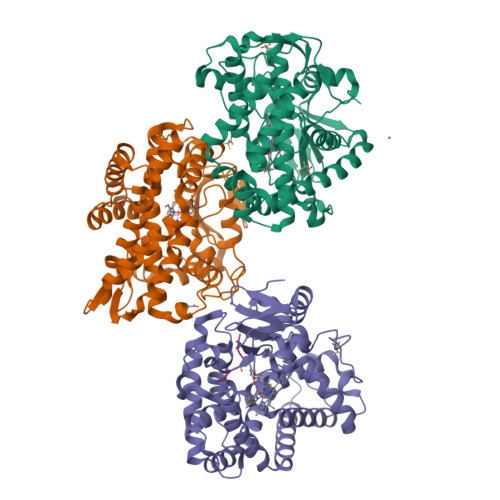
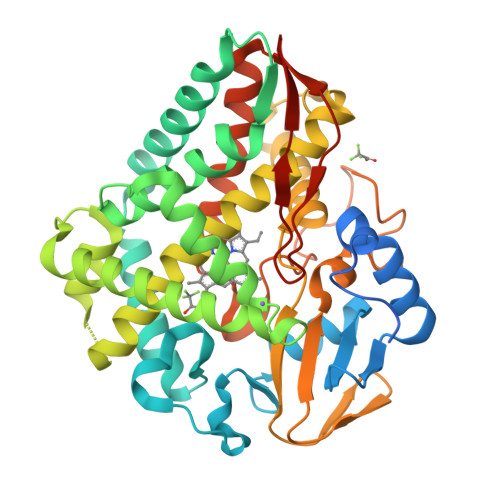
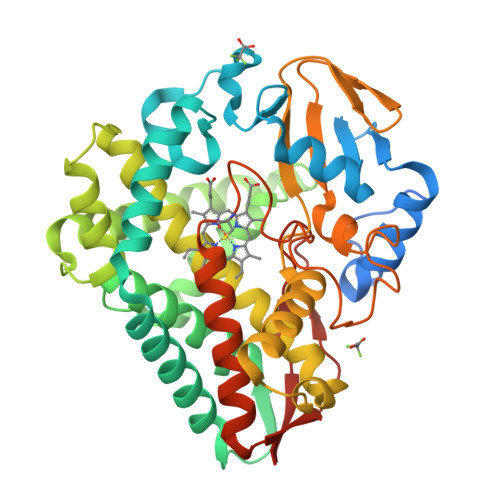
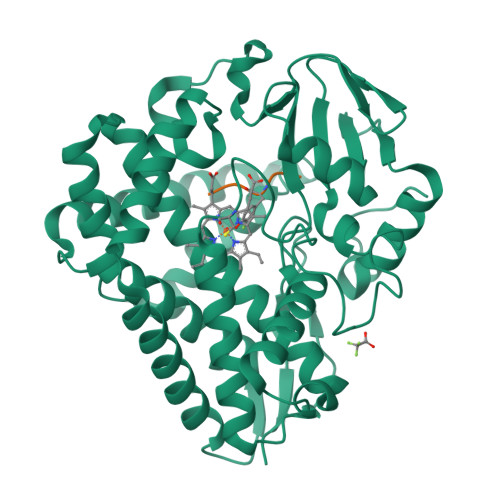
Loss of fluorine during crosslinking by the biarylitide P450 Blt proceeds due to restricted peptide orientation.
Zhao, Y., Gullick, J., Hansen, M.H., Coe, L., Treisman, M., Schittenhelm, R.B., McKay, A., Murray, L.A.M., Tailhades, J., De Voss, J.J., Krenske, E.H., Cryle, M.J.(2024) Chem Commun (Camb) 60: 13951-13954
- PubMed: 39512132
- DOI: https://doi.org/10.1039/d4cc04092a
- Primary Citation of Related Structures:
9BW8 - PubMed Abstract:
The biarylitide crosslinking enzyme P450 Blt can perform crosslinking between m -F-Tyr-3 and His-5 residues within peptide substrates with concomitant and specific loss of fluorine. Our investigations suggest that a small intrinsic preference for coupling ipso to fluorine is magnified by the binding of the peptide in a specific orientation that enforces the loss of fluorine during peptide crosslinking, likely via a two-step reaction mechanism involving the non-enzyme catalysed reductive elimination of fluoride.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The Monash Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia. max.cryle@monash.edu.