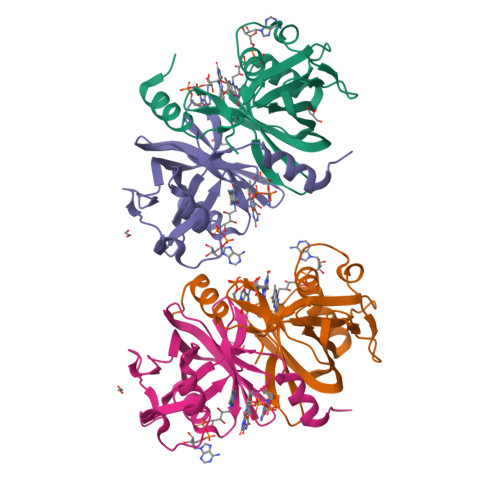
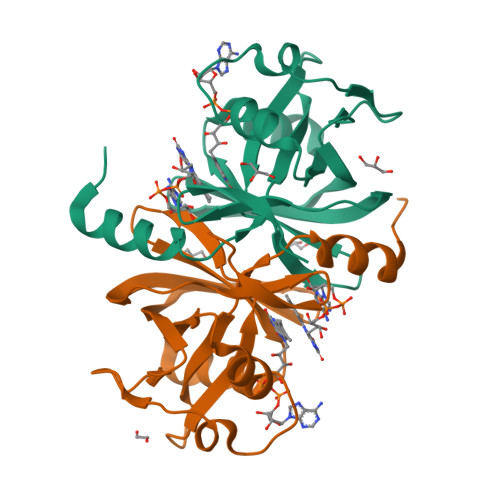
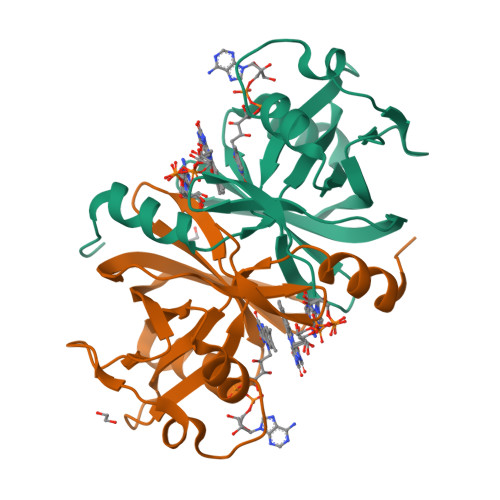
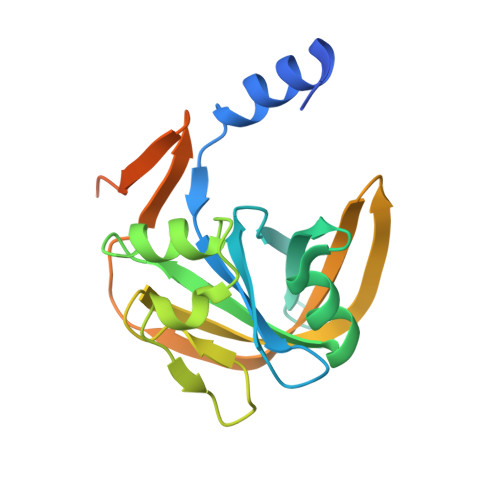
The NADH-dependent flavin reductase ThdF follows an ordered sequential mechanism though crystal structures reveal two FAD molecules in the active site.
Horstmeier, H.J., Bork, S., Nagel, M.F., Keller, W., Spross, J., Diepold, N., Ruppel, M., Kottke, T., Niemann, H.H.(2024) J Biological Chem 301: 108128-108128
- PubMed: 39725031
- DOI: https://doi.org/10.1016/j.jbc.2024.108128
- Primary Citation of Related Structures:
9FD4, 9FD5, 9FD6 - PubMed Abstract:
Two-component flavin-dependent monooxygenases are of great interest as biocatalysts for the production of pharmaceuticals and other relevant molecules, as they catalyze chemically important reactions such as hydroxylation, epoxidation and halogenation. The monooxygenase components require a separate flavin reductase, which provides the necessary reduced flavin cofactor. The tryptophan halogenase Thal from Streptomyces albogriseolus is a well-characterized two-component flavin-dependent halogenase. Thal exhibits some limitations in terms of halogenation efficiency, also caused by unproductive enzyme-substrate complexes with reduced flavin adenine dinucleotide (FAD). Since the reductase components have an important regulatory function for the activity and efficiency of the monooxygenase by controlling the supply of reduced flavin, here we studied the so far uncharacterized flavin reductase ThdF from the same gene cluster in S. albogriseolus, which potentially cooperates with Thal. A crystal structure of ThdF in complex with both substrates, FAD and NADH, revealed their orientation for hydride transfer. We obtained two further ThdF structures with two FAD molecules bound to the active site, suggesting a ping-pong bi-bi mechanism. In contrast, steady-state enzyme kinetics clearly showed that ThdF catalyzes flavin reduction via an ordered sequential mechanism, with FAD being bound first and FADH 2 released last. Compared to related flavin reductases, ThdF has a low k cat and low K M value. The inhibition of ThdF by NAD + might limit Thal's halogenation activity when the cellular NADH level is low. These results provide first insights into how the efficiency of Thal could be controlled by flavin reduction at the reductase ThdF.
Organizational Affiliation:
Structural Biochemistry, Department of Chemistry, Bielefeld University, Universitätsstraße 25, 33615 Bielefeld, Germany.