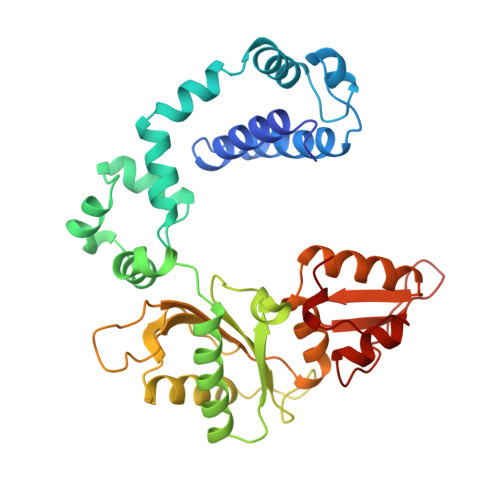
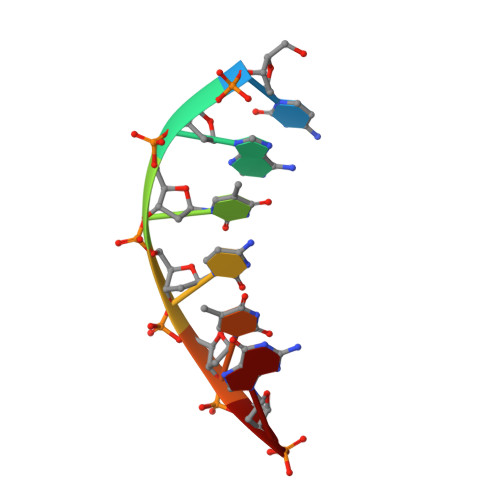
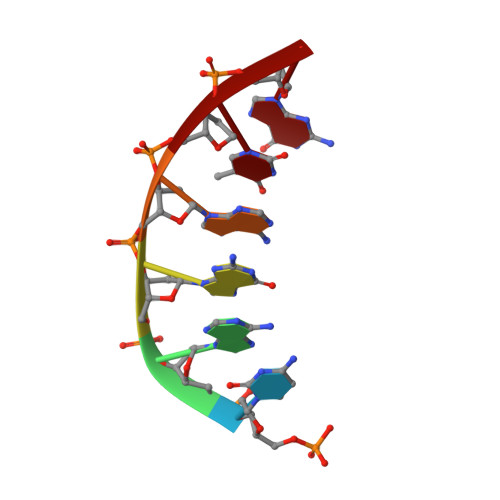
Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity
Pelletier, H., Sawaya, M.R., Wolfle, W., Wilson, S.H., Kraut, J.(1996) Biochemistry 35: 12742-12761
- PubMed: 8841118
- DOI: https://doi.org/10.1021/bi952955d
- Primary Citation of Related Structures:
9ICM, 9ICW, 9ICX, 9ICY - PubMed Abstract:
Mammalian DNA polymerase beta (pol beta) is a small (39 kDa) DNA gap-filling enzyme that comprises an amino-terminal 8-kDa domain and a carboxy-terminal 31-kDa domain. In the work reported here, crystal structures of human pol beta complexed with blunt-ended segments of DNA show that, although the crystals belong to a different space group, the DNA is nevertheless bound in the pol beta binding channel in the same way as the DNA in previously reported structures of rat pol beta complexed with a template-primer and ddCTP [Pelletier, H., Sawaya, M. R., Kumar, A., Wilson, S. H., & Kraut, J. (1994) Science 264, 1891-1903]. The 8-kDa domain is in one of three previously observed positions relative to the 31-kDa domain, suggesting that the 8-kDa domain may assume only a small number of stable conformations. The thumb subdomain is in a more open position in the human pol beta-DNA binary complex than it is in the rat pol beta-DNA-ddCTP ternary complex, and a closing thumb upon nucleotide binding could represent the rate-limiting conformational change that has been observed in pre-steady-state kinetic studies. Intermolecular contacts between the DNA and the 8-kDa domain of a symmetry-related pol beta molecule reveal a plausible binding site on the 8-kDa domain for the downstream oligonucleotide of a gapped-DNA substrate; in addition to a lysine-rich binding pocket that accommodates a 5'-PO4 end group, the 8-kDa domain also contains a newly discovered helix-hairpin-helix (HhH) motif that binds to DNA in the same way as does a structurally and sequentially homologous HhH motif in the 31-kDa domain. DNA binding by both HhH motifs is facilitated by a metal ion. In that HhH motifs have been identified in other DNA repair enzymes and DNA polymerases, the HhH-DNA interactions observed in pol beta may be applicable to a broad range of DNA binding proteins. The sequence similarity between the HhH motif of endonuclease III from Escherichia coli and the HhH motif of the 8-kDa domain of pol beta is particularly striking in that all of the conserved residues are clustered in one short sequence segment, LPGVGXK, where LPGV corresponds to a type II beta-turn (the hairpin turn), and GXK corresponds to a part of the HhH motif that is proposed to be critical for DNA binding and catalysis for both enzymes. These results suggest that endonuclease III and the 8-kDa domain of pol beta may employ a similar mode of DNA binding and may have similar catalytic mechanisms for their respective DNA lyase activities. A model for productive binding of pol beta to a gapped-DNA substrate requires a 90 degrees bend in the single-stranded template, which could enhance nucleotide selectivity during DNA repair or replication.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla 92093-0506, USA.