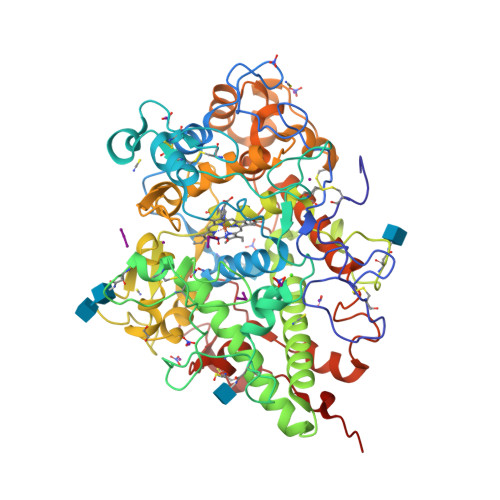
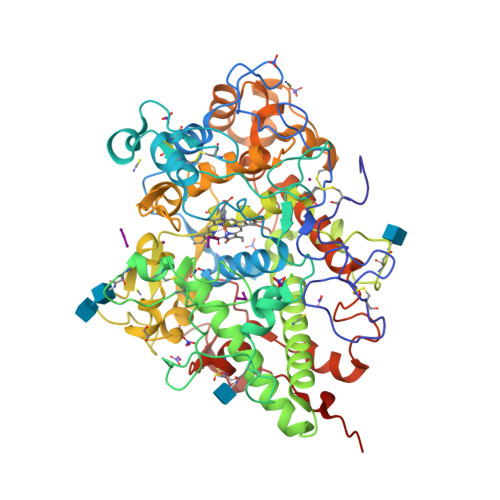
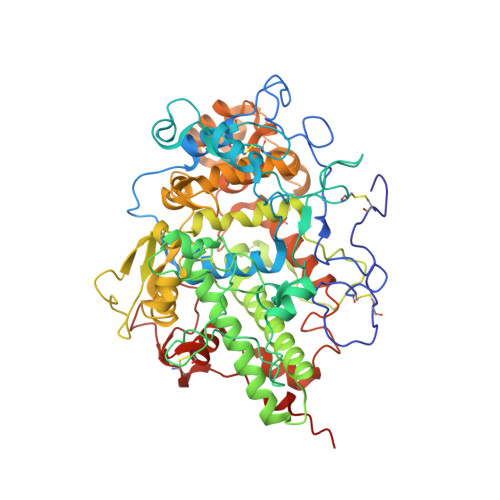
Structure of the Complex of Lactoperoxidase With Nitric Oxide at 1.95 angstrom Resolution.
Maurya, A., Ahmad, N., Sharma, P., Sharma, S., Singh, T.P.(2025) Proteins
- PubMed: 39748619
- DOI: https://doi.org/10.1002/prot.26797
- Primary Citation of Related Structures:
9IT8 - PubMed Abstract:
Lactoperoxidase (LPO) is a heme-containing mammalian enzyme that is found in the extracellular fluids of animals including plasma, saliva, airway epithelial and nasal lining fluids, milk, tears, and gastric juices. LPO uses hydrogen peroxide (H 2 O 2 ) to convert substrates into oxidized products. Previous structural studies have shown that H 2 O 2 , CO, and CN are bound to LPO at the distal heme cavity by coordinating with heme iron. The structure of the complex of LPO with NO shows that NO also binds to LPO at the distal heme cavity and forms a coordinate linkage with heme iron. The structure shows that the nitrogen atom of NO is linked to heme iron at a distance of 1.97 while the oxygen atom is attached to the N ε2 atom of His109 at a distance of 2.23 Å. On the other hand, N atom of NO is located with an interatomic distance of 3.25 Å allowing a hydrogen-bonding interaction with the N ε2 atom of Gln105. A comparison of the bindings of NO, CO, CN, and H 2 O 2 in coordination with heme iron indicates stereochemical compatibility of the distal heme cavity for the binding of diatomic molecules. However, notable differences are observed in their orientations in the distal heme cavity indicating functional differences. The bindings of NO, CO, and CN by coordinating with heme iron result in the inhibition of LPO while the binding of H 2 O 2 to heme iron produces an intermediate of LPO known as Compound I.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.