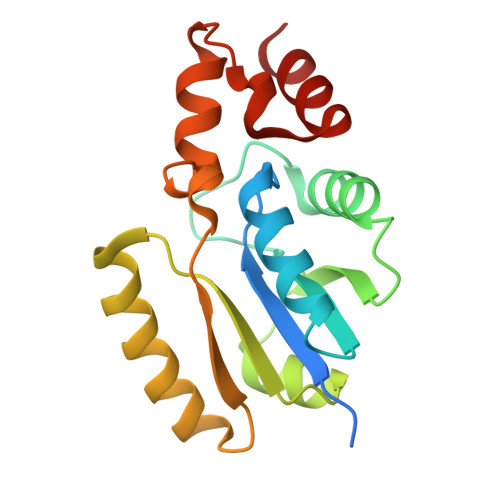
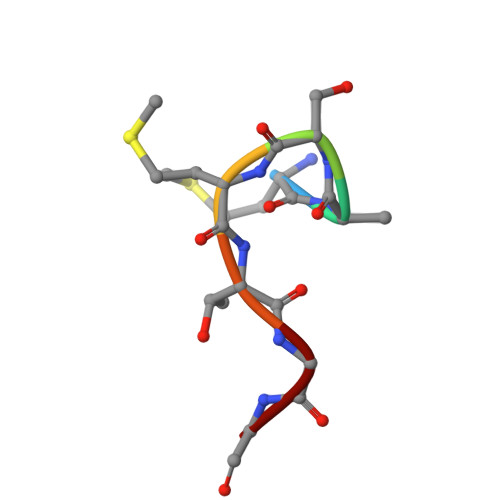
Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.20 A resolution.
Ahmad, N., Sharma, P., Sharma, S., Singh, T.P.To be published.