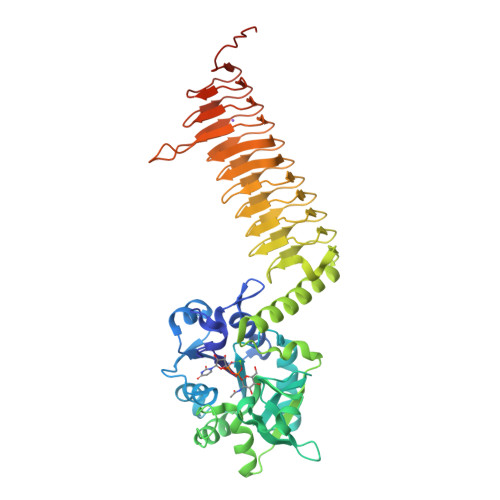
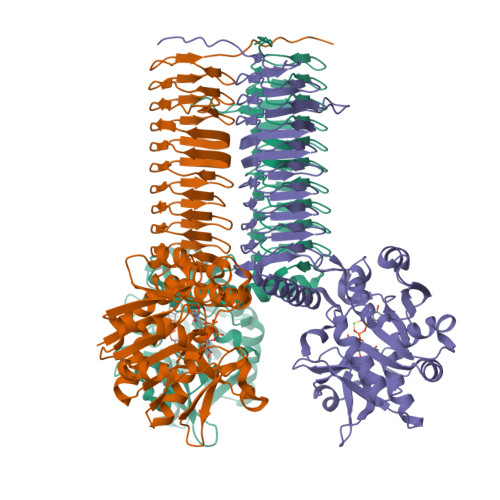
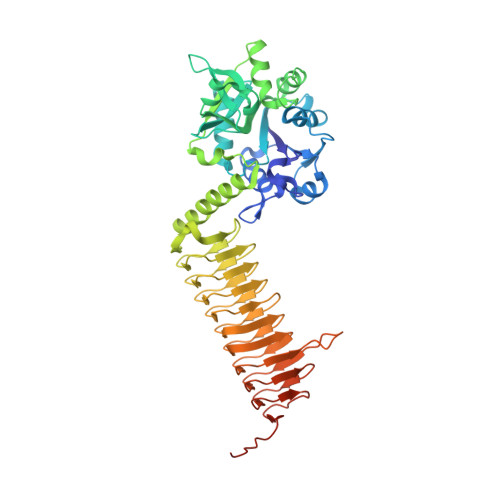
Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.
Kostrewa, D., D'Arcy, A., Takacs, B., Kamber, M.(2001) J Mol Biology 305: 279-289
- PubMed: 11124906
- DOI: https://doi.org/10.1006/jmbi.2000.4296
- Primary Citation of Related Structures:
1G95, 1G97 - PubMed Abstract:
N-Acetylglucosamine-1-phosphate uridyltransferase (GlmU) is an essential bacterial enzyme with both an acetyltransferase and a uridyltransferase activity which have been mapped to the C-terminal and N-terminal domains, respectively. GlmU performs the last two steps in the synthesis of UDP-N-acetylglucosamine (UDP-GlcNAc), which is an essential precursor in both the peptidoglycan and the lipopolysaccharide metabolic pathways. GlmU is therefore an attractive target for potential antibiotics. Knowledge of its three-dimensional structure would provide a basis for rational drug design. We have determined the crystal structures of Streptococcus pneumoniae GlmU (SpGlmU) in apo form at 2.33 A resolution, and in complex with UDP-N-acetyl glucosamine and the essential co-factor Mg(2+) at 1.96 A resolution. The protein structure consists of an N-terminal domain with an alpha/beta-fold, containing the uridyltransferase active site, and a C-terminal domain with a long left-handed beta-sheet helix (LbetaH) domain. An insertion loop containing the highly conserved sequence motif Asn-Tyr-Asp-Gly protrudes from the left-handed beta-sheet helix domain. In the crystal, S. pneumoniae GlmU forms exact trimers, mainly through contacts between left-handed beta-sheet helix domains. UDP-N-acetylglucosamine and Mg(2+) are bound at the uridyltransferase active site, which is in a closed form. We propose a uridyltransferase mechanism in which the activation energy of the double negatively charged phosphorane transition state is lowered by charge compensation of Mg(2+) and the side-chain of Lys22.
Organizational Affiliation:
Pharmaceutical Research Chemical Technologies, F. Hoffmann-La Roche Ltd., Basle, 4070, Switzerland. dirk.kostrewa@psi.ch