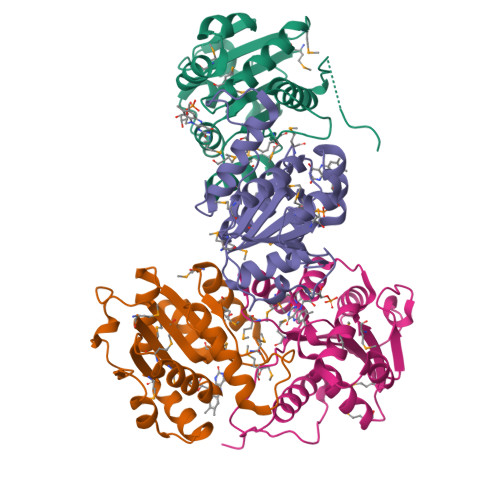
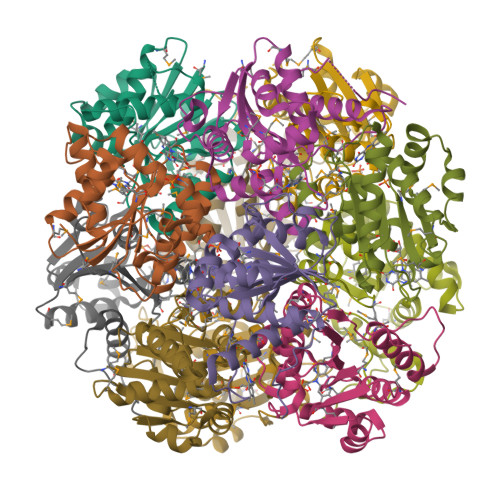
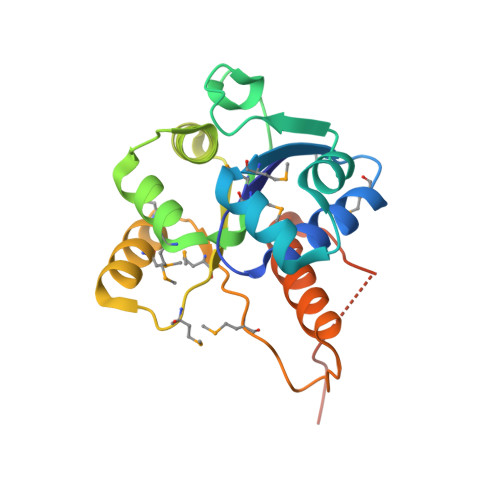
Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Rangarajan, E.S., Li, Y., Iannuzzi, P., Tocilj, A., Hung, L.-W., Matte, A., Cygler, M.(2004) Protein Sci 13: 3006-3016
- PubMed: 15459342
- DOI: https://doi.org/10.1110/ps.04953004
- Primary Citation of Related Structures:
1SBZ - PubMed Abstract:
The crystal structure of the flavoprotein Pad1 from Escherichia coli O157:H7 complexed with the cofactor FMN has been determined by the multiple anomalous diffraction method and refined at 2.0 A resolution. This protein is a paralog of UbiX (3-octaprenyl-4-hydroxybenzoate carboxylyase, 51% sequence identity) that catalyzes the third step in ubiquinone biosynthesis and to Saccharomyces cerevisiae Pad1 (54% identity), an enzyme that confers resistance to the antimicrobial compounds phenylacrylic acids through decarboxylation of these compounds. Each Pad1 monomer consists of a typical Rossmann fold containing a non-covalently bound molecule of FMN. The fold of Pad1 is similar to MrsD, an enzyme associated with lantibiotic synthesis; EpiD, a peptidyl-cysteine decarboxylase; and AtHAL3a, the enzyme, which decarboxylates 4'-phosphopantothenoylcysteine to 4'-phosphopantetheine during coenzyme A biosynthesis, all with a similar location of the FMN binding site at the interface between two monomers, yet each having little sequence similarity to one another. All of these proteins associate into oligomers, with a trimer forming the common structural unit in each case. In MrsD and EpiD, which belong to the homo-dodecameric flavin-containing cysteine decarboxylase (HFCD) family, these trimers associate further into dodecamers. Pad1 also forms dodecamers, although the association of the trimers is completely different, resulting in exposure of a different side of the trimer unit to the solvent. This exposure affects the location of the substrate binding site and, specifically, its access to the FMN cofactor. Therefore, Pad1 forms a separate family, distinguishable from the HFCD family.
Organizational Affiliation:
Biotechnology Research Institute, NRCC, 6100 Royalmount Avenue, Montreal, Quebec H4P 2R2, Canada.