Novel zinc finger motif in the basal transcriptional machinery: three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS.
Qian, X., Gozani, S.N., Yoon, H., Jeon, C.J., Agarwal, K., Weiss, M.A.(1993) Biochemistry 32: 9944-9959
- PubMed: 8399164
- DOI: https://doi.org/10.1021/bi00089a010
- Primary Citation of Related Structures:
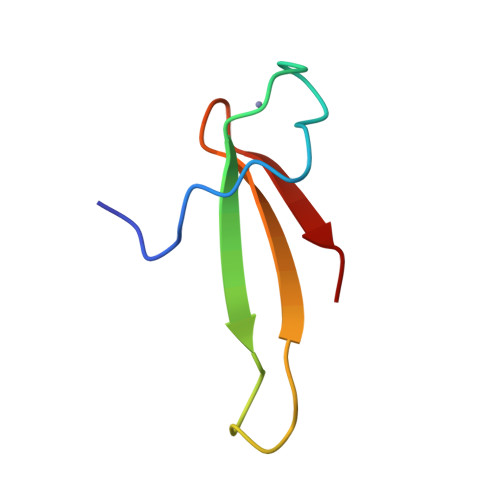
1TFI - PubMed Abstract:
Transcriptional elongation provides a key control point in the regulation of eukaryotic gene expression. Here we describe homonuclear and 15N-heteronuclear 3D NMR studies of the nucleic acid binding domain of human transcriptional elongation factor TFIIS. This domain contains a Cys4 Zn(2+)-binding site with no homology to previously characterized Cys4, Cys6, or Cys2-His2 Zn fingers. Complete 1H and 15N NMR resonance assignment of a 50-residue TFIIS peptide-Zn2+ complex is obtained. Its solution structure, as determined by distance geometry/simulated annealing (DG/SA) calculations, exhibits a novel three-stranded antiparallel beta-sheet (designated the Zn ribbon). Analogous sequence motifs occur in a wide class of proteins involved in RNA or DNA transactions, including human basal transcriptional initiation factor TFIIE. A three-dimensional model of the TFIIE Cys4 domain is obtained by DG-based homology modeling. The role of the TFIIS Zn ribbon in the control of eukaryotic transcriptional elongation is discussed.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115.