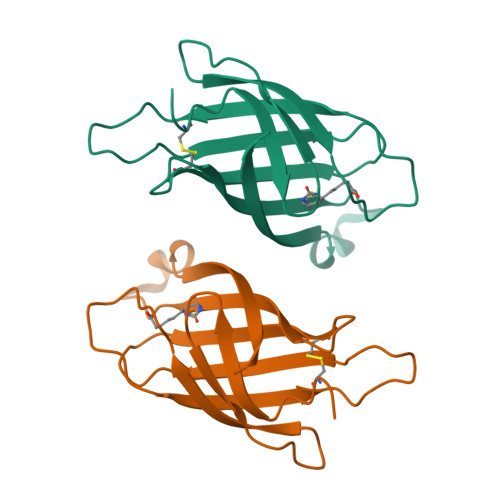
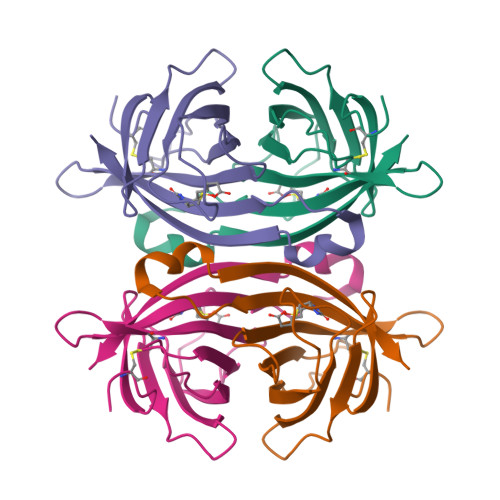
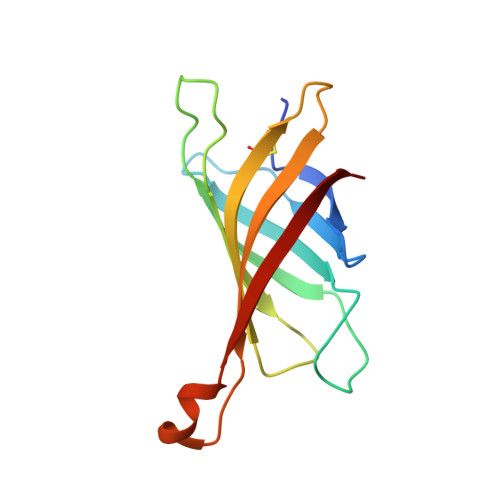
Structure and Characterization of a Novel Chicken Biotin-Binding Protein a (Bbp-A).
Hytonen, V.P., Maatta, J.A.E., Niskanen, E.A., Huuskonen, J., Helttunen, K.J., Halling, K.K., Nordlund, H.R., Rissanen, K., Johnson, M.S., Salminen, T.A., Kulomaa, M.S., Laitinen, O.H., Airenne, T.T.(2007) BMC Struct Biol 7: 8
- PubMed: 17343730
- DOI: https://doi.org/10.1186/1472-6807-7-8
- Primary Citation of Related Structures:
2C1Q, 2C1S - PubMed Abstract:
The chicken genome contains a BBP-A gene showing similar characteristics to avidin family genes. In a previous study we reported that the BBP-A gene may encode a biotin-binding protein due to the high sequence similarity with chicken avidin, especially at regions encoding residues known to be located at the ligand-binding site of avidin. Here, we expand the repertoire of known macromolecular biotin binders by reporting a novel biotin-binding protein A (BBP-A) from chicken. The BBP-A recombinant protein was expressed using two different expression systems and purified with affinity chromatography, biochemically characterized and two X-ray structures were solved - in complex with D-biotin (BTN) and in complex with D-biotin D-sulfoxide (BSO). The BBP-A protein binds free biotin with high, "streptavidin-like" affinity (Kd ~ 10-13 M), which is about 50 times lower than that of chicken avidin. Surprisingly, the affinity of BBP-A for BSO is even higher than the affinity for BTN. Furthermore, the solved structures of the BBP-A--BTN and BBP-A--BSO complexes, which share the fold with the members of the avidin and lipocalin protein families, are extremely similar to each other. BBP-A is an avidin-like protein having a beta-barrel fold and high affinity towards BTN. However, BBP-A differs from the other known members of the avidin protein family in thermal stability and immunological properties. BBP-A also has a unique ligand-binding property, the ability to bind BTN and BSO at comparable affinities. BBP-A may have use as a novel material in, e.g. modern bio(nano)technological applications.
Organizational Affiliation:
NanoScience Center, Department of Biological and Environmental Science, P.O. Box 35 (YAB), FI-40014 University of Jyväskylä, Finland. veshyto@mat.ethz.ch