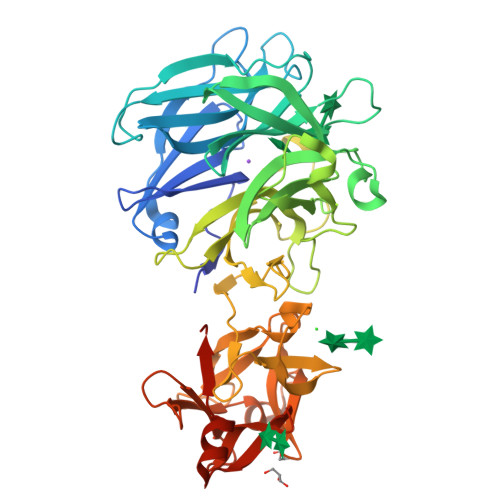
Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
Fujimoto, Z., Ichinose, H., Maehara, T., Honda, M., Kitaoka, M., Kaneko, S.(2010) J Biological Chem 285: 34134-34143
- PubMed: 20739278
- DOI: https://doi.org/10.1074/jbc.M110.164251
- Primary Citation of Related Structures:
3AKF, 3AKG, 3AKH, 3AKI - PubMed Abstract:
Exo-1,5-α-L-arabinofuranosidases belonging to glycoside hydrolase family 43 have strict substrate specificity. These enzymes hydrolyze only the α-1,5-linkages of linear arabinan and arabino-oligosaccharides in an exo-acting manner. The enzyme from Streptomyces avermitilis contains a core catalytic domain belonging to glycoside hydrolase family 43 and a C-terminal arabinan binding module belonging to carbohydrate binding module family 42. We determined the crystal structure of intact exo-1,5-α-L-arabinofuranosidase. The catalytic module is composed of a 5-bladed β-propeller topologically identical to the other family 43 enzymes. The arabinan binding module had three similar subdomains assembled against one another around a pseudo-3-fold axis, forming a β-trefoil-fold. A sugar complex structure with α-1,5-L-arabinofuranotriose revealed three subsites in the catalytic domain, and a sugar complex structure with α-L-arabinofuranosyl azide revealed three arabinose-binding sites in the carbohydrate binding module. A mutagenesis study revealed that substrate specificity was regulated by residues Asn-159, Tyr-192, and Leu-289 located at the aglycon side of the substrate-binding pocket. The exo-acting manner of the enzyme was attributed to the strict pocket structure of subsite -1, formed by the flexible loop region Tyr-281-Arg-294 and the side chain of Tyr-40, which occupied the positions corresponding to the catalytic glycon cleft of GH43 endo-acting enzymes.
Organizational Affiliation:
Protein Research Unit, National Institute of Agrobiological Sciences, 2-1-2 Kannondai, Tsukuba, Ibaraki 305-8602, Japan.