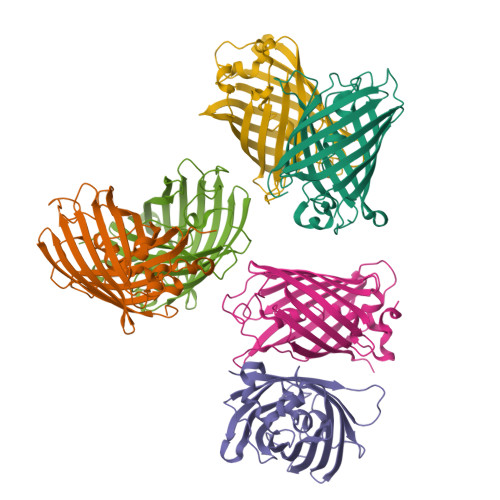
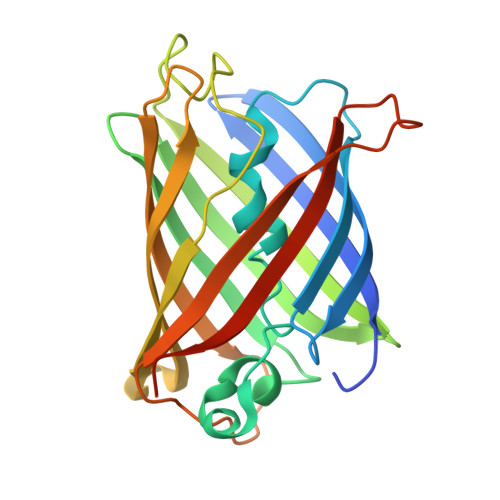
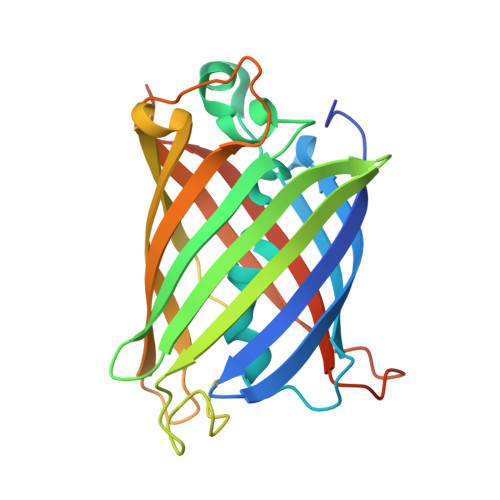
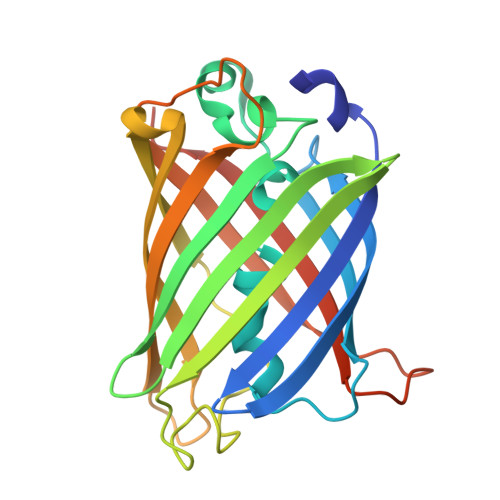
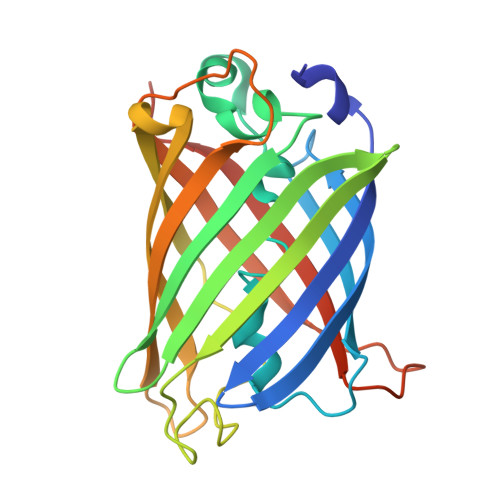
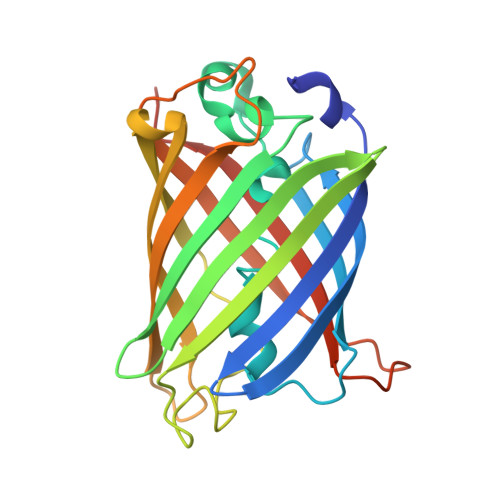
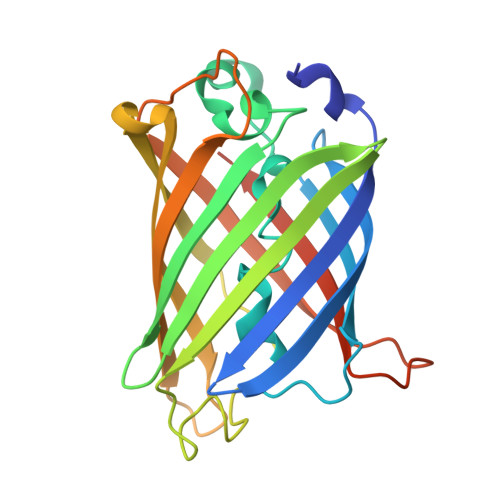
The mechanism of folding robustness revealed by the crystal structure of extra-superfolder GFP.
Choi, J.Y., Jang, T.-H., Park, H.H.(2017) FEBS Lett 591: 442-447
- PubMed: 27990640
- DOI: https://doi.org/10.1002/1873-3468.12534
- Primary Citation of Related Structures:
5B61 - PubMed Abstract:
Stability of green fluorescent protein (GFP) is sometimes important for a proper practical application of this protein. Random mutagenesis and targeted mutagenesis have been used to create better-folded variants of GFP, including recently reported extra-superfolder GFP. Our aim was to determine the crystal structure of extra-superfolder GFP, which is more robustly folded and stable than GFP and superfolder GFP. The structural and structure-based mutagenesis analyses revealed that some of the mutations that created extra-superfolder GFP (F46L, E126K, N149K, and S208L) contribute to folding robustness by stabilizing extra-superfolder GFP with various noncovalent bonds.
Organizational Affiliation:
School of Biotechnology and Graduate School of Biochemistry, Yeungnam University, Gyeongsan, South Korea.