Facile Fabrication of Protein-Macrocycle Frameworks.
Ramberg, K.O., Engilberge, S., Skorek, T., Crowley, P.B.(2021) J Am Chem Soc 143: 1896-1907
- PubMed: 33470808
- DOI: https://doi.org/10.1021/jacs.0c10697
- Primary Citation of Related Structures:
6Z5G, 6Z5M, 6Z5P, 6Z5Q, 6Z5W, 6Z5X, 6Z5Z, 6Z60, 6Z62, 7ALF, 7ALG - PubMed Abstract:
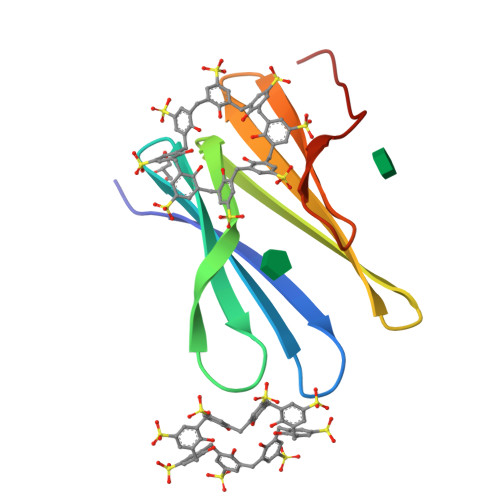
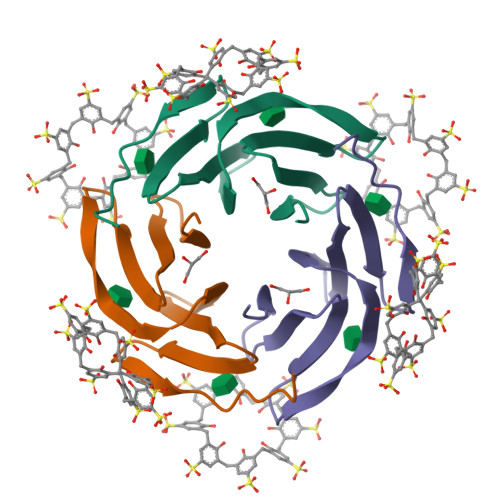
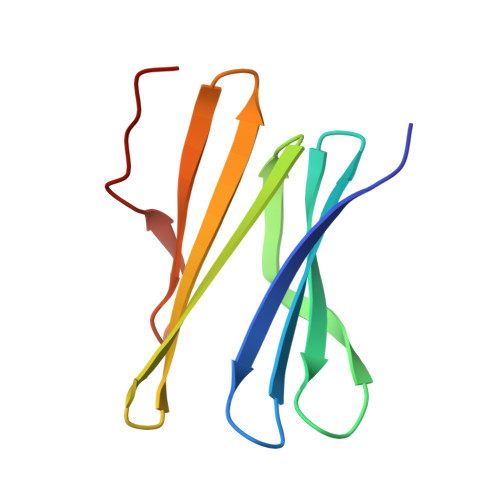
Precisely defined protein aggregates, as exemplified by crystals, have applications in functional materials. Consequently, engineered protein assembly is a rapidly growing field. Anionic calix[n]arenes are useful scaffolds that can mold to cationic proteins and induce oligomerization and assembly. Here, we describe protein-calixarene composites obtained via cocrystallization of commercially available sulfonato-calix[8]arene ( sclx 8 ) with the symmetric and "neutral" protein RSL. Cocrystallization occurred across a wide range of conditions and protein charge states, from pH 2.2-9.5, resulting in three crystal forms. Cationization of the protein surface at pH ∼ 4 drives calixarene complexation and yielded two types of porous frameworks with pore diameters >3 nm. Both types of framework provide evidence of protein encapsulation by the calixarene. Calixarene-masked proteins act as nodes within the frameworks, displaying octahedral-type coordination in one case. The other framework formed millimeter-scale crystals within hours, without the need for precipitants or specialized equipment. NMR experiments revealed macrocycle - modulated side chain p K a values and suggested a mechanism for pH-triggered assembly. The same low pH framework was generated at high pH with a permanently cationic arginine-enriched RSL variant. Finally, in addition to protein framework fabrication, sclx 8 enables de novo structure determination.
Organizational Affiliation:
School of Chemistry, National University of Ireland Galway, University Road, Galway, H91 TK33, Ireland.