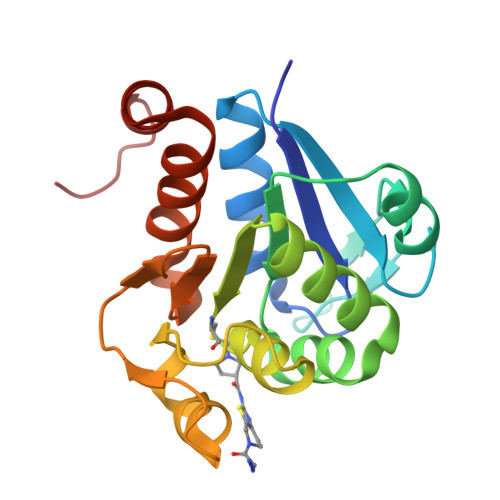
Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
Jia, Y., Kim, R.Q., Kooij, R., Ovaa, H., Sapmaz, A., Geurink, P.P.(2022) J Med Chem 65: 13288-13304
- PubMed: 36149939
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01113
- Primary Citation of Related Structures:
7PA2, 7PA3 - PubMed Abstract:
The multifunctional human Parkinson's disease protein 7 (PARK7/DJ1) is an attractive therapeutic target due to its link with early-onset Parkinson's disease, upregulation in various cancers, and contribution to chemoresistance. However, only a few compounds have been identified to bind PARK7 due to the lack of a dedicated chemical toolbox. We report the creation of such a toolbox and showcase the application of each of its components. The selective PARK7 submicromolar inhibitor with a cyanimide reactive group covalently modifies the active site Cys106. Installment of different dyes onto the inhibitor delivered two PARK7 probes. The Rhodamine110 probe provides a high-throughput screening compatible FP assay, showcased by screening a compound library (8000 molecules). The SulfoCy5-equipped probe is a valuable tool to assess the effect of PARK7 inhibitors in a cell lysate. Our work creates new possibilities to explore PARK7 function in a physiologically relevant setting and develop new and improved PARK7 inhibitors.
Organizational Affiliation:
Oncode Institute & Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, Leiden 2333 ZC, The Netherlands.