A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme.
Furuya, T., Nakane, D., Kitanishi, K., Katsuumi, N., Tsaturyan, A., Shcherbakov, I.N., Unno, M., Akitsu, T.(2023) Sci Rep 13: 6892-6892
- PubMed: 37106030
- DOI: https://doi.org/10.1038/s41598-023-33926-1
- Primary Citation of Related Structures:
7YRK - PubMed Abstract:
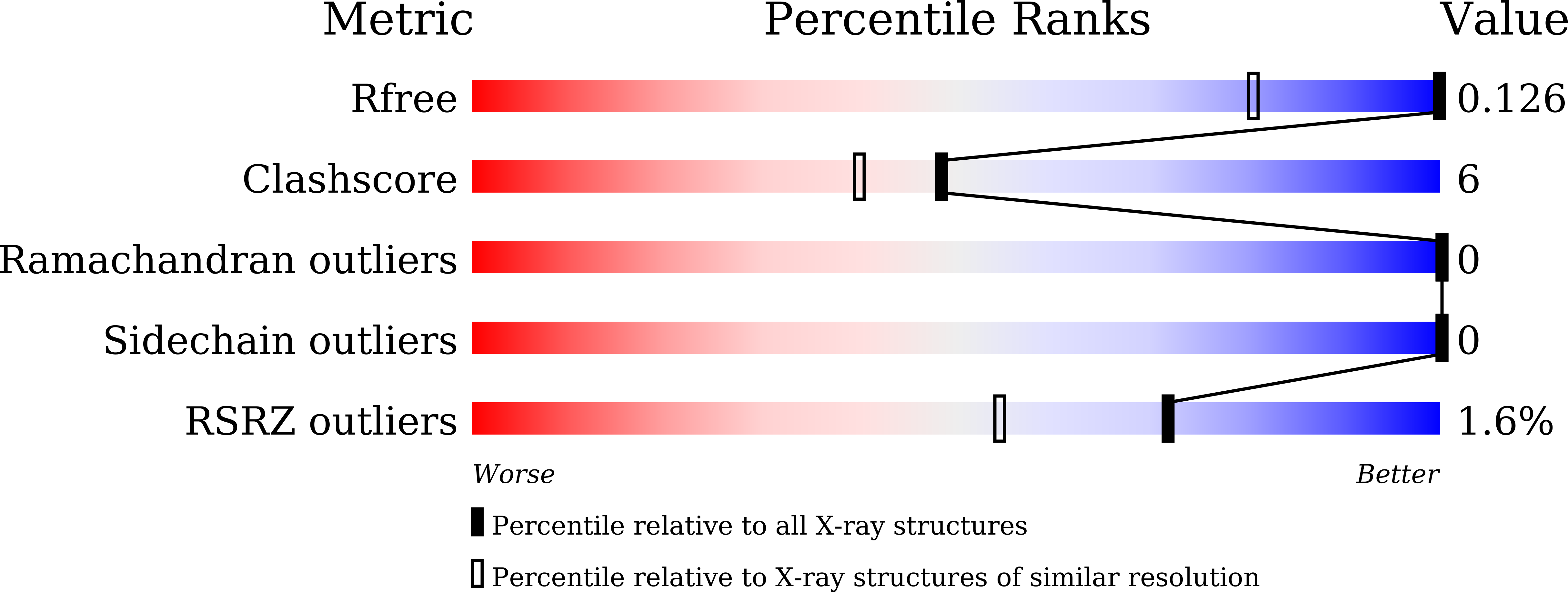
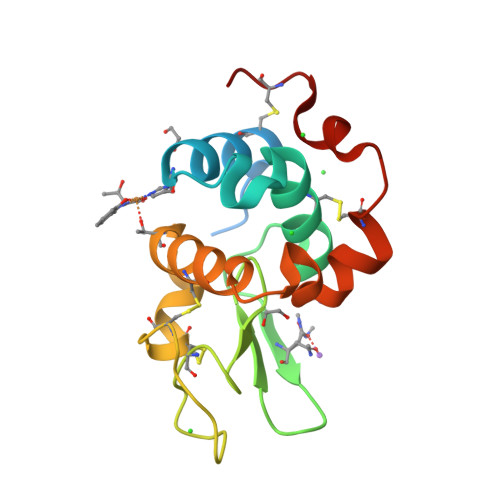
A novel hybrid protein composed of a superoxide dismutase-active Cu(II) complex (CuST) and lysozyme (CuST@lysozyme) was prepared. The results of the spectroscopic and electrochemical analyses confirmed that CuST binds to lysozyme. We determined the crystal structure of CuST@lysozyme at 0.92 Å resolution, which revealed that the His15 imidazole group of lysozyme binds to the Cu(II) center of CuST in the equatorial position. In addition, CuST was fixed in position by the weak axial coordination of the Thr89 hydroxyl group and the hydrogen bond between the guanidinium group of the Arg14 residue and the hydroxyl group of CuST. Furthermore, the combination of CuST with lysozyme did not decrease the superoxide dismutase activity of CuST. Based on the spectral, electrochemical, structural studies, and quantum chemical calculations, an O 2 - disproportionation mechanism catalyzed by CuST@lysozyme is proposed.
Organizational Affiliation:
Department of Chemistry, Faculty of Science, Tokyo University of Science, 1-3 Kagurazaka, Shinjuku-ku, Tokyo, 162-8601, Japan.