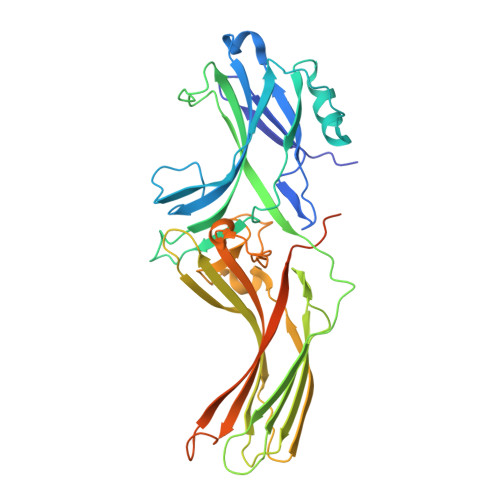
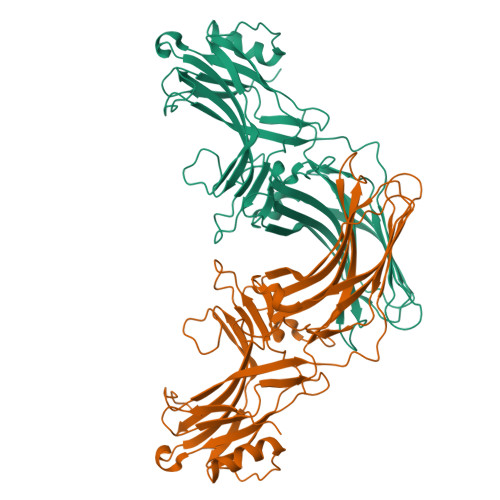
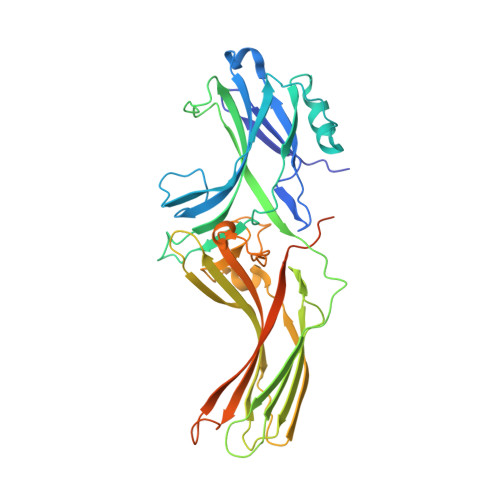
Mechanisms of amphibian arrestin 1 self-association and dynamic distribution in retinal photoreceptors.
Barnes, C.L., Salom, D., Namitz, K.E.W., Smith, W.C., Knutson, B.A., Cosgrove, M.S., Kiser, P.D., Calvert, P.D.(2024) J Biological Chem 300: 107966-107966
- PubMed: 39510183
- DOI: https://doi.org/10.1016/j.jbc.2024.107966
- Primary Citation of Related Structures:
8FUT, 8FUU - PubMed Abstract:
Visual arrestin 1 (Arr1) is an essential protein for termination of the light response in photoreceptors. While mammalian Arr1s form dimers and tetramers at physiological concentrations in vitro, oligomerization in other vertebrates has not been studied. Here we examine self-association of Arr1 from two amphibian species, Xenopus laevis (xArr1) and Ambystoma tigrinum (salArr1). Sedimentation velocity analytical ultracentrifugation showed that xArr1 and salArr1 oligomerization is limited to dimers. The K D for dimer formation was 53 μM for xArr1 and 44 μM for salArr1, similar to the 69 μM K D for bovine Arr1 (bArr1) dimers. Mutations of orthologous amino acids important for mammalian Arr1 oligomerization had no impact on xArr1 dimerization. Crystallography showed that the fold of xArr1 closely resembles that of bArr1 and crystal structures in different space groups revealed two potential xArr1 dimer forms: a symmetric dimer with a C-domain interface (CC dimer), resembling the bArr1 solution dimer, and an asymmetric dimer with an N-domain/C-domain interface. Mutagenesis of residues predicted to interact in either of these two dimer forms yielded modest reduction in dimer affinity, suggesting that the dimer interfaces compete or are not unique. Indeed, small-angle X-ray scattering and protein painting data were consistent with a symmetric anti-parallel solution dimer (AP dimer) distinct from the assemblies observed by crystallography. Finally, a computational model evaluating xArr1 binding to compartment-specific partners and partitioning based on heterogeneity of available cytoplasmic spaces shows that Arr1 distribution in dark-adapted photoreceptors is largely explained by the excluded volume effect together with tuning by oligomerization.
Organizational Affiliation:
Center for Vision Research and the Department of Ophthalmology and Visual Sciences, SUNY Upstate Medical University, Syracuse, New York, USA.