Reconstitution of human DNA licensing and the structural and functional analysis of key intermediates.
Wells, J.N., Edwardes, L.V., Leber, V., Allyjaun, S., Peach, M., Tomkins, J., Kefala-Stavridi, A., Faull, S.V., Aramayo, R., Pestana, C.M., Ranjha, L., Speck, C.(2025) Nat Commun 16: 478-478
- PubMed: 39779677
- DOI: https://doi.org/10.1038/s41467-024-55772-z
- Primary Citation of Related Structures:
8RWV - PubMed Abstract:
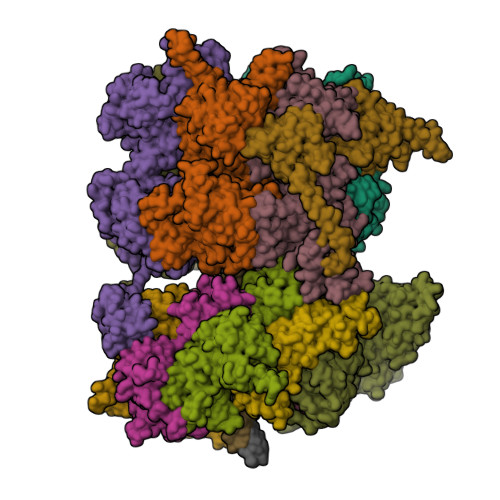
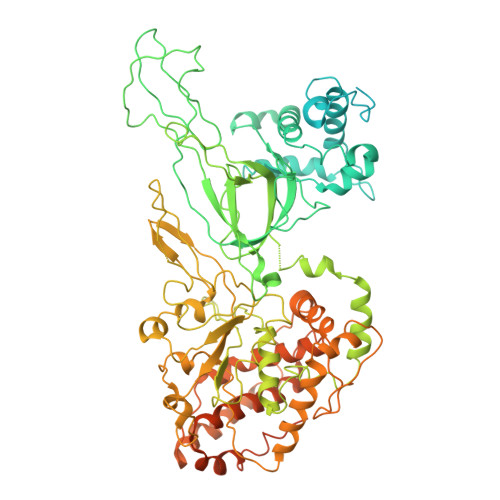
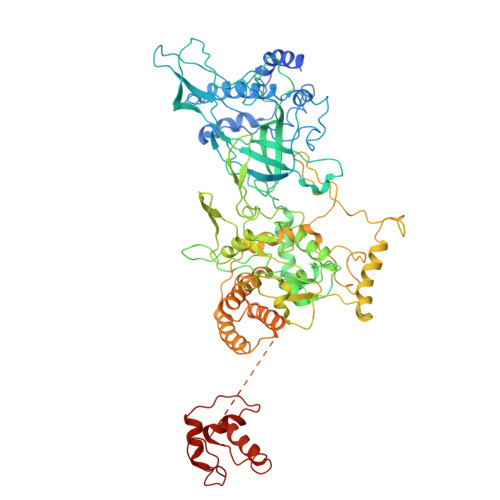
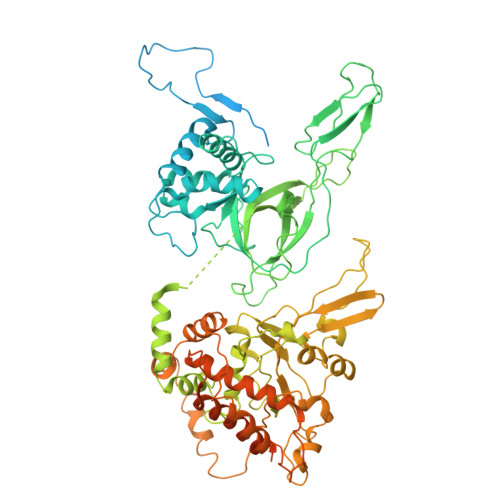
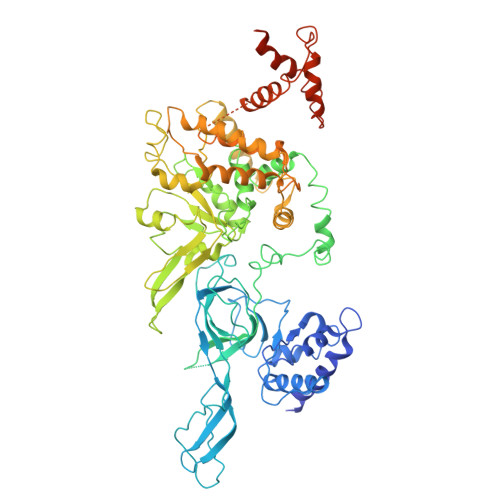
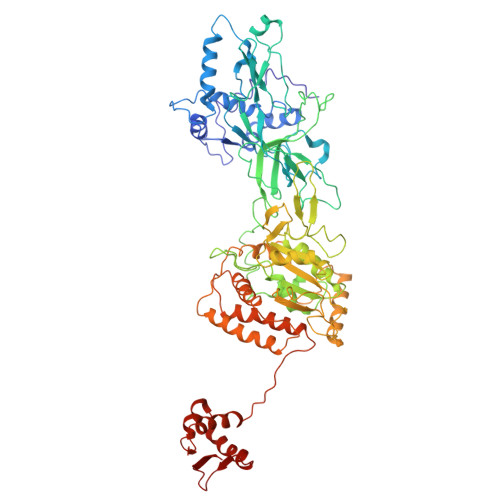
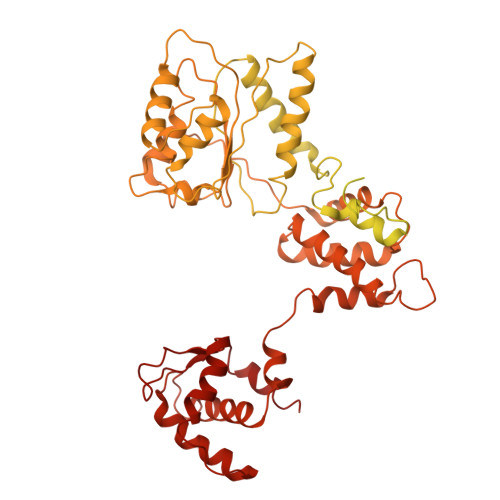
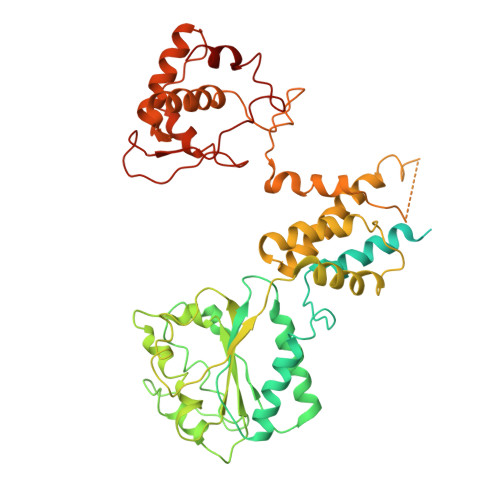
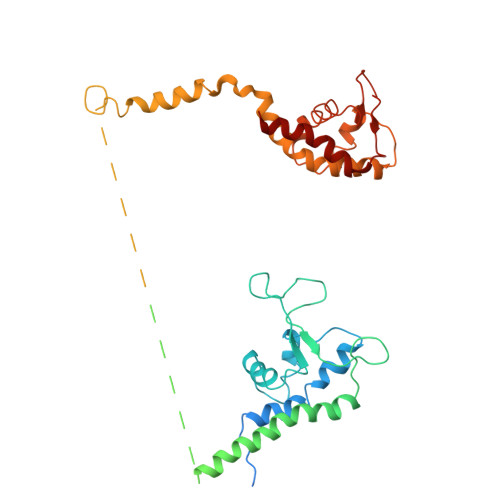
Human DNA licensing initiates replication fork assembly and DNA replication. This reaction promotes the loading of the hMCM2-7 complex on DNA, which represents the core of the replicative helicase that unwinds DNA during S-phase. Here, we report the reconstitution of human DNA licensing using purified proteins. We showed that the in vitro reaction is specific and results in the assembly of high-salt resistant hMCM2-7 double-hexamers. With ATPγS, an hORC1-5-hCDC6-hCDT1-hMCM2-7 (hOCCM) assembles independent of hORC6, but hORC6 enhances double-hexamer formation. We determined the hOCCM structure, which showed that hORC-hCDC6 recruits hMCM2-7 via five hMCM winged-helix domains. The structure highlights how hORC1 activates the hCDC6 ATPase and uncovered an unexpected role for hCDC6 ATPase in complex disassembly. We identified that hCDC6 binding to hORC1-5 stabilises hORC2-DNA interactions and supports hMCM3-dependent recruitment of hMCM2-7. Finally, the structure allowed us to locate cancer-associated mutations at the hCDC6-hMCM3 interface, which showed specific helicase loading defects.
Organizational Affiliation:
DNA Replication Group, Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, UK.