NMR Solution Structure of the 2:1 Coptisine-ATF4-G4 Complex
Xiao, C.M., Li, Y.P., Liu, Y.S., Dong, R.F., He, X.Y., Lin, Q., Zang, X., Wang, K.B., Xia, Y.Z., Kong, L.Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*TP*AP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*AP*GP*GP*GP*AP*GP*C*(LIG)*(LIG))-3') | A [auth X] | 22 | Homo sapiens | 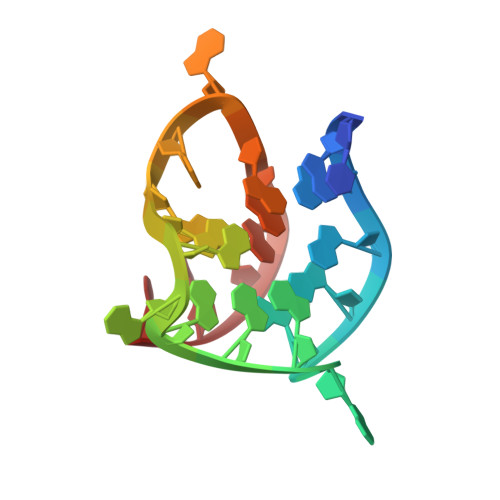 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| KPT (Subject of Investigation/LOI) Query on KPT | B [auth X], C [auth X] | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium C19 H14 N O4 XYHOBCMEDLZUMP-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 82173707 |