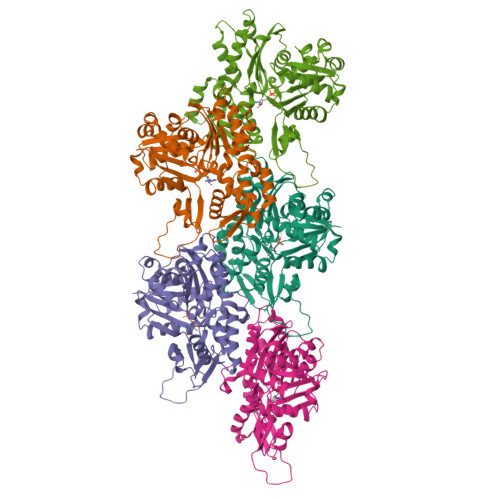
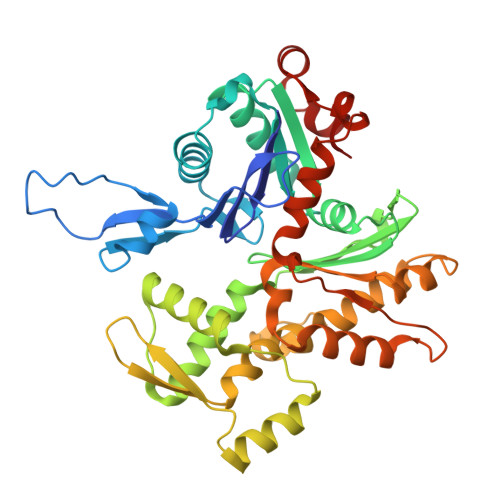
Cryo-EM reconstruction of yeast ADP-actin filament at 2.5 angstrom resolution. A comparison with vertebrate F-actin.
Stevenson, S.R., Tzokov, S.B., Lahiri, I., Ayscough, K.R., Bullough, P.A.(2025) Structure 33: 435-442.e3
- PubMed: 39798573
- DOI: https://doi.org/10.1016/j.str.2024.12.008
- Primary Citation of Related Structures:
9GO5 - PubMed Abstract:
The core component of the actin cytoskeleton is the globular protein G-actin, which reversibly polymerizes into filaments (F-actin). Budding yeast possesses a single actin that shares 87%-89% sequence identity with vertebrate actin isoforms. Previous structural studies indicate very close overlap of main-chain backbones. Intriguingly, however, substitution of yeast ACT1 with vertebrate β-cytoplasmic actin severely disrupts cell function and the substitution with a skeletal muscle isoform is lethal. Here we report a 2.5 Å structure of budding yeast F-actin. Previously unresolved side-chain information allows us to highlight four main differences in the comparison of yeast and vertebrate ADP F-actins: a more open nucleotide binding pocket; a more solvent exposed C-terminus; a rearrangement of inter-subunit binding interactions in the vicinity of the D loop and changes in the hydrogen bonding network in the vicinity of histidine 73 (yeast actin) and methyl-histidine 73 (vertebrate actin).
Organizational Affiliation:
Molecular and Cell Biology, School of Biosciences, University of Sheffield, Sheffield S10 2TN, UK.