Full Text |
QUERY: Gene Name = "YLR442C" | MyPDB Login | Search API |
| Search Summary | This query matches 18 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 18 of 18 Structures Page 1 of 1 Sort by
Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3Zhou, Z., Feng, H., Zhou, B., Ghirlando, R., Hu, K., Zwolak, A., Jenkins, L., Xiao, H., Tjandra, N., Wu, C., Bai, Y. (2011) Nature 472: 234-237
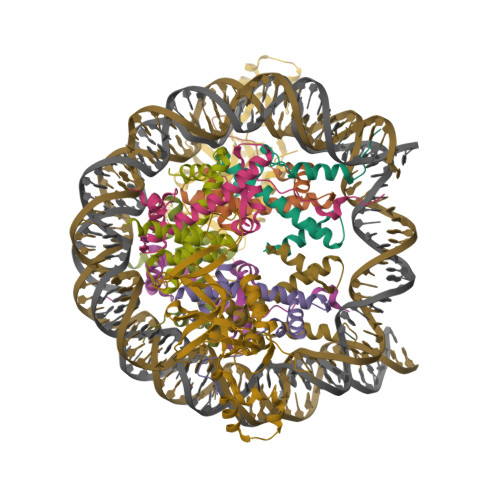
Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosomeWang, F., Li, G., Mohammed, A., Lu, C., Currie, M., Johnson, A., Moazed, D. (2013) Proc Natl Acad Sci U S A 110: 8495-8500
Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particleYang, D., Fang, Q., Wang, M., Ren, R., Wang, H., He, M., Sun, Y., Yang, N., Xu, R.M. (2013) Nat Struct Mol Biol 20: 1116-1118
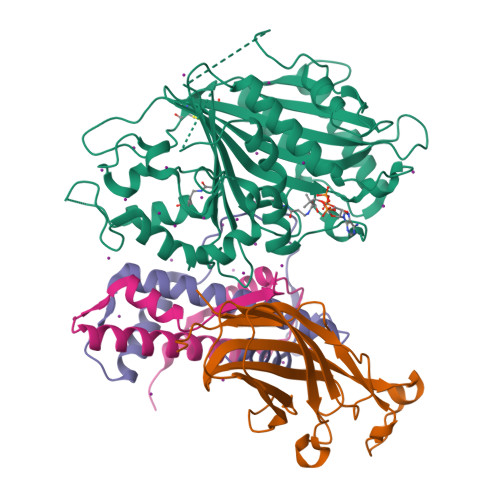
Crystal structure of histone acetyltransferase complex(2014) Genes Dev 28: 1217-1227
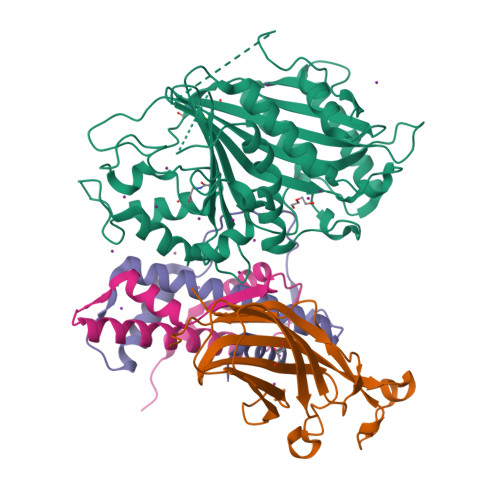
Crystal structure of Rtt109-Asf1-H3-H4-CoA complexZhang, L., Serra-Cardona, A., Zhou, H., Wang, M., Yang, N., Zhang, Z., Xu, R.M. (2018) Cell 174: 818-830.e11
Crystal structure of Rtt109-Asf1-H3-H4 complexZhang, L., Serra-Cardona, A., Zhou, H., Wang, M., Yang, N., Zhang, Z., Xu, R.M. (2018) Cell 174: 818-830.e11
Chromatin remodeller-nucleosome complex at 3.6 A resolution.Willhoft, O., Chua, E.Y.D., Wilkinson, M., Wigley, D.B. (2018) Science 362:
Chromatin remodeller-nucleosome complex at 4.5 A resolution.Willhoft, O., Chua, E.Y.D., Wilkinson, M., Wigley, D.B. (2018) Science 362:
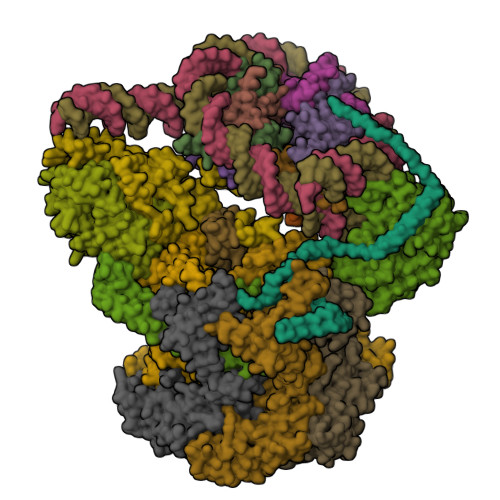
Structure of inner kinetochore CCAN-Cenp-A complexYan, K., Yang, J., Zhang, Z., McLaughlin, S.H., Chang, L., Fasci, D., Heck, A.J.R., Barford, D. (2019) Nature 574: 278-282
Cryo-EM structure of the 1:1 Orc1 BAH domain in complex with nucleosomeJiang, H., Yu, C., Liu, C.P., Han, X., Yu, Z., Xu, R.M. To be published
Cryo-EM structure of the 2:1 Orc1 BAH domain in complex with nucleosomeJiang, H., Yu, C., Liu, C.P., Han, X., Yu, Z., Xu, R.M. To be published
antibody and nucleosome complex(2021) Nat Commun 12: 1763-1763
nucleosome and Gal4 complex(2021) Nat Commun 12: 1763-1763
Crystal structure of Hat1-Hat2-Asf1-H3-H4(2022) Genes Dev 36: 408-413
Structure of transcription factor UAF in complex with TBP and 35S rRNA promoter DNABaudin, F., Murciano, B., Fung, H.K.H., Fromm, S.A., Mueller, C.W. (2022) Sci Adv 8: eabn5725-eabn5725
Composite model of the yeast Hir Complex with Asf1/H3/H4(2024) Mol Cell 84: 2601-2617.e12
Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosomeDendooven, T.D., Zhang, Z., Yang, J., McLaughlin, S., Schwabb, J., Scheres, S., Yatskevich, S., Barford, D. (2023) Sci Adv 9: eadg7480-eadg7480
Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome.Dendooven, T.D., Zhang, Z., Yang, J., McLaughlin, S., Schwabb, J., Scheres, S., Yatskevich, S., Barford, D. (2023) Sci Adv 9: eadg7480-eadg7480
1 to 18 of 18 Structures Page 1 of 1 Sort by |