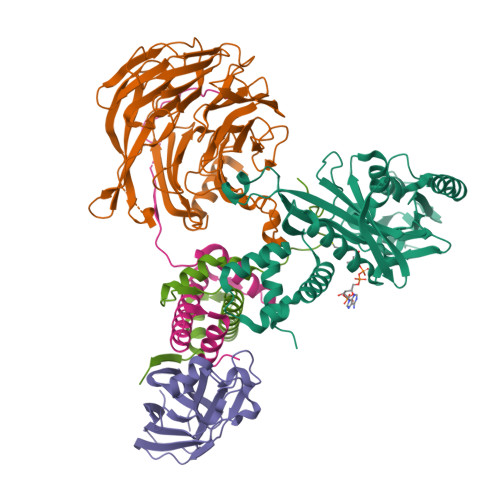
Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Yue, Y., Yang, W.S., Zhang, L., Liu, C.P., Xu, R.M.(2022) Genes Dev 36: 408-413
- PubMed: 35393344
- DOI: https://doi.org/10.1101/gad.349099.121
- Primary Citation of Related Structures:
7XAY - PubMed Abstract:
Chaperones influence histone conformation and intermolecular interaction in multiprotein complexes, and the structures obtained with full-length histones often provide more accurate and comprehensive views. Here, our structure of the Hat1-Hat2 acetyltransferase complex bound to Asf1-H3-H4 shows that the core domains of H3 and H4 are involved in binding Hat1 and Hat2, and the N-terminal tail of H3 makes extensive interaction with Hat2. These findings expand the knowledge about histone-protein interaction and implicate a function of Hat2/RbAp46/48, which is a versatile histone chaperone found in many chromatin-associated complexes, in the passing of histones between chaperones.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.