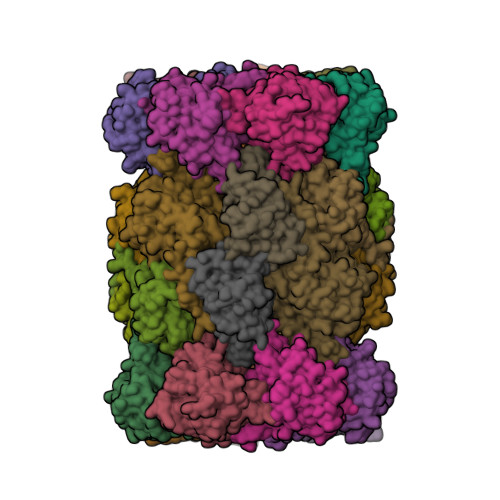
Structure Determination MethodologyScientific Name of Source OrganismRefinement Resolution (Å)Enzyme Classification Name | Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.7 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 6N5, CL, MES, MG |
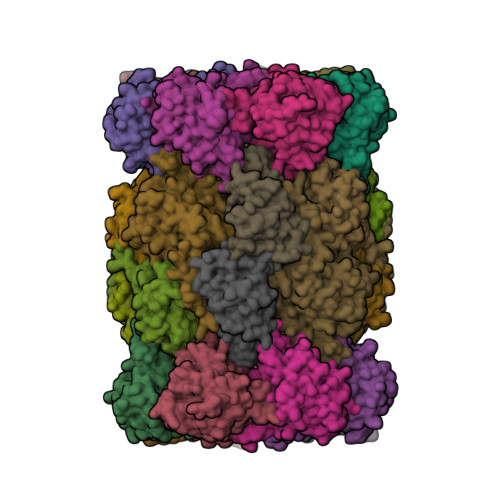
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.8 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 79P, CL, MES, MG |
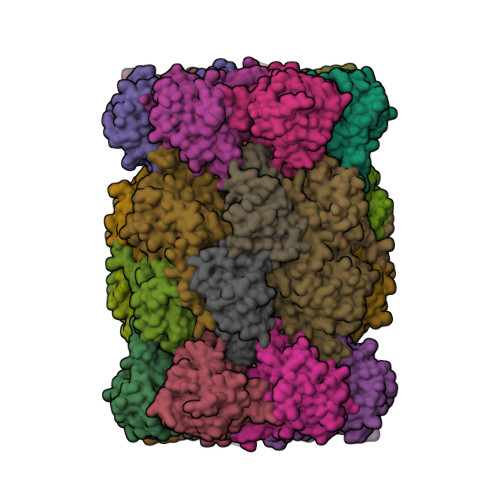
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.9 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 6NV, CL, MES, MG |
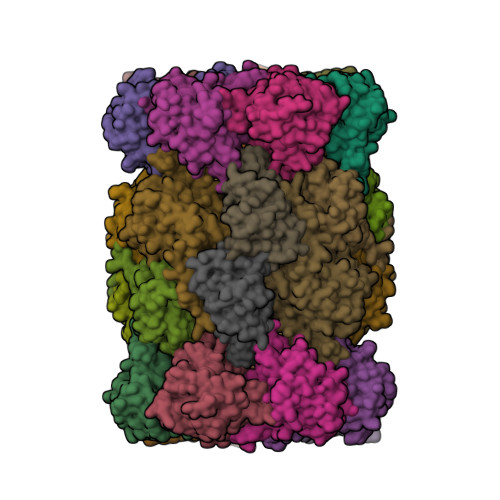
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.4 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | CL, MG |
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.8 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | CL, MG |
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.8 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 04C, CL, MES, MG |
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.9 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 3BV, CL, MES, MG |
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.5 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | BO2, CL, MG |
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.6 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 39V, CL, MES, MG |
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.9 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 38X, CL, MES, MG |
Groll, M., Huber, E.M. (2016) EMBO J 35: 2602-2613 | Released | 2016-11-09 | | Method | X-RAY DIFFRACTION 2.9 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | 6N5, CL, MES, MG |
de la Pena, A.H., Goodall, E.A., Gates, S.N., Lander, G.C., Martin, A. (2018) Science 362: | Released | 2018-10-17 | | Method | ELECTRON MICROSCOPY 4.43 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | ADP, ATP |
de la Pena, A.H., Goodall, E.A., Gates, S.N., Lander, G.C., Martin, A. (2018) Science 362: | Released | 2018-10-17 | | Method | ELECTRON MICROSCOPY 4.73 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | ADP, ATP |
de la Pena, A.H., Goodall, E.A., Gates, S.N., Lander, G.C., Martin, A. (2018) Science 362: | Released | 2018-10-17 | | Method | ELECTRON MICROSCOPY 4.27 Å | | Organisms | | | Macromolecule | Unique protein chains: 14 | | Unique Ligands | ADP, ATP |
de la Pena, A.H., Goodall, E.A., Gates, S.N., Lander, G.C., Martin, A. (2018) Science 362: | Released | 2018-10-17 | | Method | ELECTRON MICROSCOPY 4.17 Å | | Organisms | | | Macromolecule | Unique protein chains: 24 | | Unique Ligands | ADP, ATP |
|