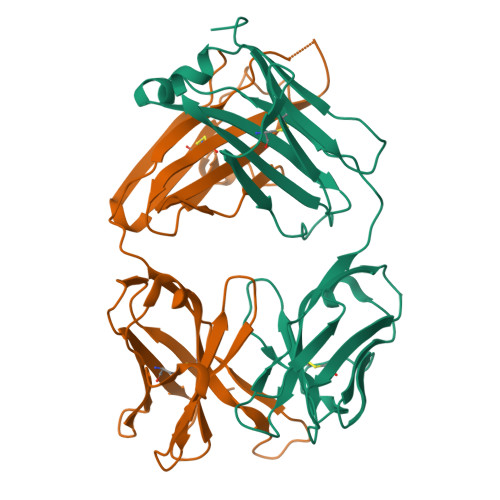
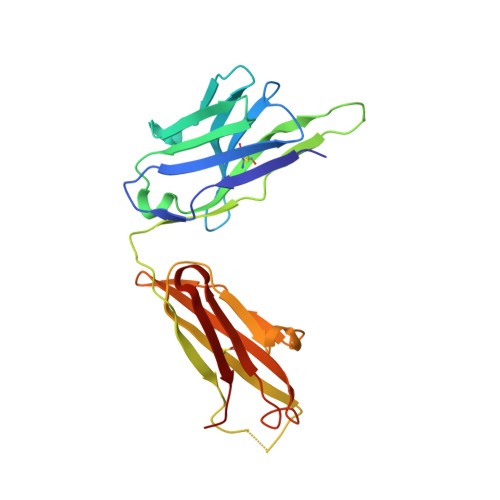
Structure of Fab hGR-2 F6, a competitive antagonist of the glucagon receptor.
Wright, L.M., Brzozowski, A.M., Hubbard, R.E., Pike, A.C., Roberts, S.M., Skovgaard, R.N., Svendsen, I., Vissing, H., Bywater, R.P.(2000) Acta Crystallogr D Biol Crystallogr 56: 573-580
- PubMed: 10771426
- DOI: https://doi.org/10.1107/s090744490000233x
- Primary Citation of Related Structures:
1DQD - PubMed Abstract:
The monoclonal antibody hGR-2 F6 has been raised against the human glucagon receptor and shown to act as a competitive antagonist. As a first step in the structural characterization of the receptor, the crystal structure of the Fab fragment from this antibody is reported at 2.1 A resolution. The hGR-2 F6 Fab crystallizes in the orthorhombic space group P2(1)2(1)2, with unit-cell parameters a = 76.14, b = 133.74, c = 37.46 A. A model generated by homology modelling was used as an aid in the chain-tracing and the Fab fragment structure was subsequently refined (final R factor = 21.7%). The structure obtained exhibits the typical immunoglobulin fold. Complementarity-determining regions (CDRs) L1, L2, L3, H1 and H2 could be superposed onto standard canonical CDR loops. The H3 loop could be classified according to recently published rules regarding loop length, sequence and conformation. This loop is 14 residues long, with an approximate beta-hairpin geometry, which is distorted somewhat by the presence of two trans proline residues at the beginning of the loop. It is expected that this H3 loop will facilitate the design of synthetic probes for the glucagon receptor that may be used to investigate receptor activity.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, University of York, York YO10 5DD, England.