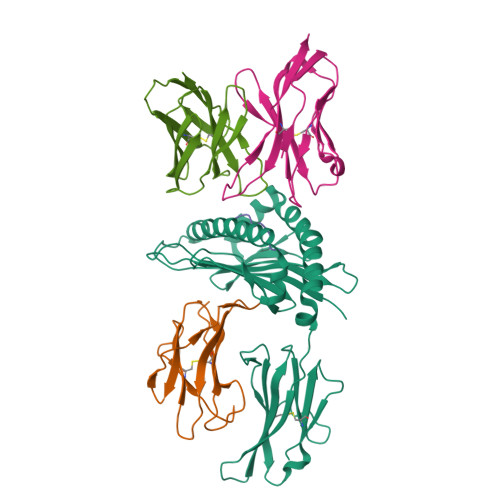
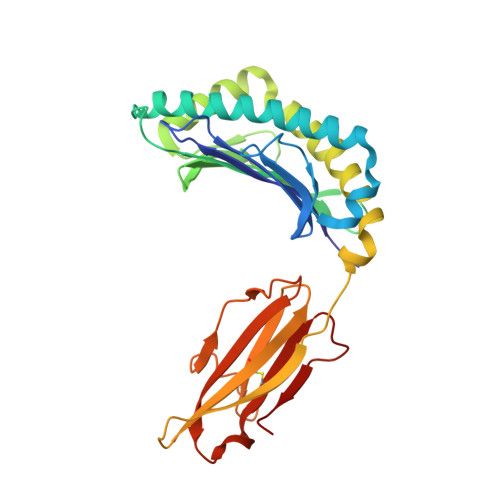
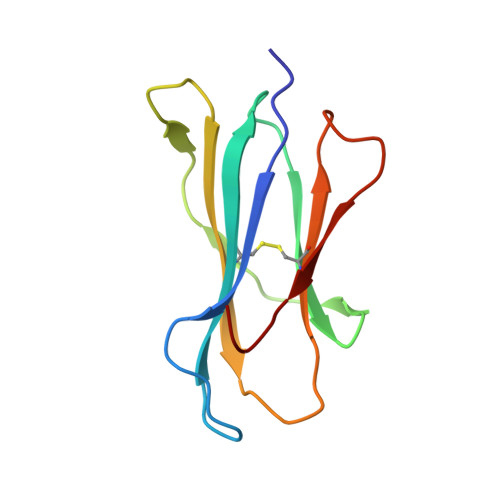
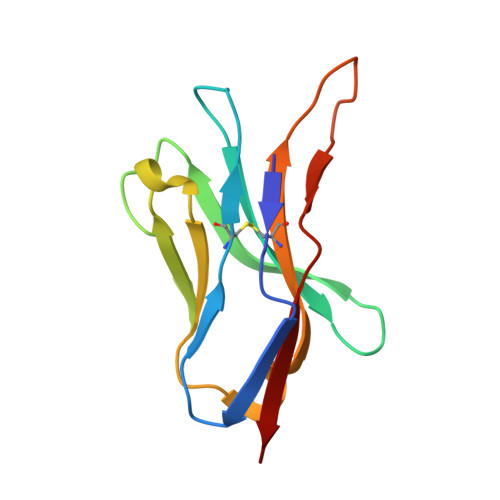
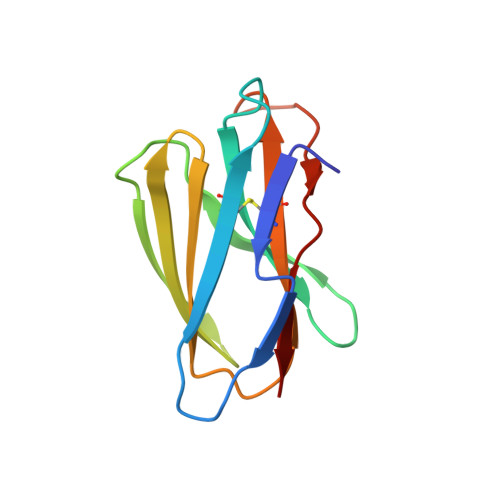
Crystal structure of a T cell receptor bound to an allogeneic MHC molecule.
Reiser, J.B., Darnault, C., Guimezanes, A., Gregoire, C., Mosser, T., Schmitt-Verhulst, A.-M., Fontecilla-Camps, J.C., Malissen, B., Housset, D., Mazza, G.(2000) Nat Immunol 1: 291-297
- PubMed: 11017099
- DOI: https://doi.org/10.1038/79728
- Primary Citation of Related Structures:
1FO0 - PubMed Abstract:
Many T cell receptors (TCRs) that are selected to respond to foreign peptide antigens bound to self major histocompatibility complex (MHC) molecules are also reactive with allelic variants of self-MHC molecules. This property, termed alloreactivity, causes graft rejection and graft-versus-host disease. The structural features of alloreactivity have yet to be defined. We now present a basis for this cross-reactivity, elucidated by the crystal structure of a complex involving the BM3.3 TCR and a naturally processed octapeptide bound to the H-2Kb allogeneic MHC class I molecule. A distinguishing feature of this complex is that the eleven-residue-long complementarity-determining region 3 (CDR3) found in the BM3.3 TCR alpha chain folds away from the peptide binding groove and makes no contact with the bound peptide, the latter being exclusively contacted by the BM3.3 CDR3 beta. Our results formally establish that peptide-specific, alloreactive TCRs interact with allo-MHC in a register similar to the one they use to contact self-MHC molecules.
Organizational Affiliation:
Laboratoire de Cristallographie et Cristallogénèse des Protéines, Institut de Biologie Structurale J.-P. Ebel, CEA-CNRS-UJF, 41, rue Jules Horowitz, F-38027 Grenoble, France.