Structural Basis for Specificity of Oxime Based Inhibitors Towards Type II Dehydroquinase from Mycobacterium Tuberculosis
Robinson, D.A., Roszak, A.W., Frederickson, M., Abell, C., Coggins, J.R., Lapthorn, A.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3-DEHYDROQUINATE DEHYDRATASE | 146 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 EC: 4.2.1.10 | 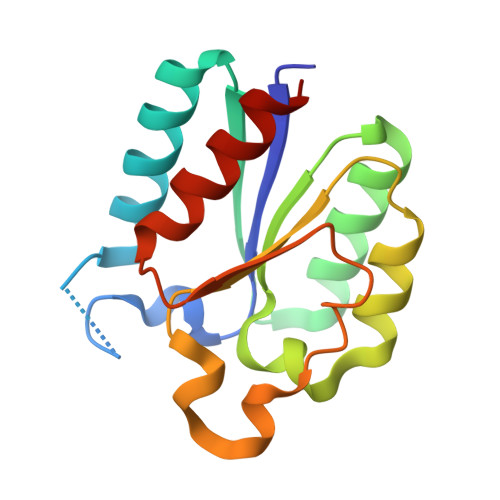 | |
UniProt | |||||
Find proteins for P9WPX7 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Go to UniProtKB: P9WPX7 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FA6 Query on FA6 | B [auth A] | 3-HYDROXYIMINO QUINIC ACID C7 H11 N O6 WASIBXJFRXJWAR-GKQOBJDDSA-N |  | ||
| SO4 Query on SO4 | C [auth A], D [auth A], E [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | F [auth A], G [auth A], H [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 126.856 | α = 90 |
| b = 126.856 | β = 90 |
| c = 126.856 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |