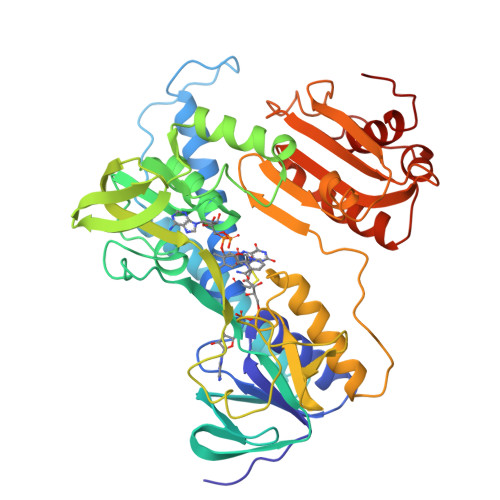
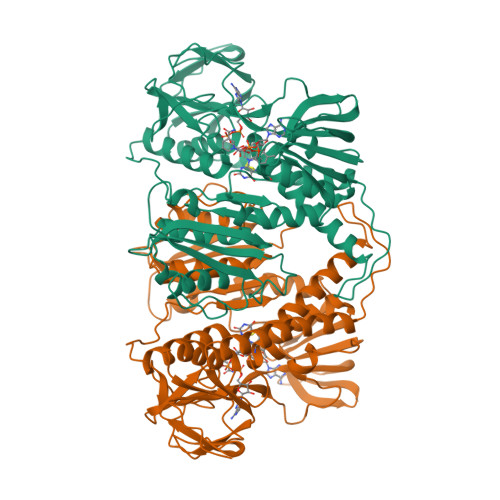
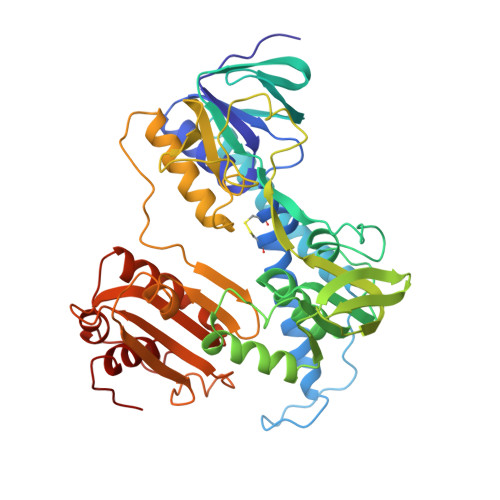
The refined crystal structure of Pseudomonas putida lipoamide dehydrogenase complexed with NAD+ at 2.45 A resolution.
Mattevi, A., Obmolova, G., Sokatch, J.R., Betzel, C., Hol, W.G.(1992) Proteins 13: 336-351
- PubMed: 1325638
- DOI: https://doi.org/10.1002/prot.340130406
- Primary Citation of Related Structures:
1LVL - PubMed Abstract:
The three-dimensional structure of one of the three lipoamide dehydrogenases occurring in Pseudomonas putida, LipDH Val, has been determined at 2.45 A resolution. The orthorhombic crystals, grown in the presence of 20 mM NAD+, contain 458 residues per asymmetric unit. A crystallographic 2-fold axis generates the dimer which is observed in solution. The final crystallographic R-factor is 21.8% for 18,216 unique reflections and a model consisting of 3,452 protein atoms, 189 solvent molecules and 44 NAD+ atoms, while the overall B-factor is unusually high: 47 A2. The structure of LipDH Val reveals the conformation of the C-terminal residues which fold "back" into the putative lipoamide binding region. The C-terminus has been proven to be important for activity by site-directed mutagenesis. However, the distance of the C-terminus to the catalytically essential residues is surprisingly large, over 6 A, and the precise role of the C-terminus still needs to be elucidated. In this crystal form LipDH Val contains one NAD+ molecule per subunit. Its adenine-ribose moiety occupies an analogous position as in the structure of glutathione reductase. However, the nicotinamide-ribose moiety is far removed from its expected position near the isoalloxazine ring and points into solution. Comparison of LipDH Val with Azotobacter vinelandii lipoamide dehydrogenase yields an rms difference of 1.6 A for 440 well defined C alpha atoms per subunit. Comparing LipDH Val with glutathione reductase shows large differences in the tertiary and quaternary structure of the two enzymes. For instance, the two subunits in the dimer are shifted by 6 A with respect to each other. So, LipDH Val confirms the surprising differences in molecular architecture between glutathione reductase and lipoamide dehydrogenase, which were already observed in Azotobacter vinelandii LipDH. This is the more remarkable since the active sites are located at the subunit interface and are virtually identical in all three enzymes.
Organizational Affiliation:
Department of Chemistry, University of Groningen, The Netherlands.