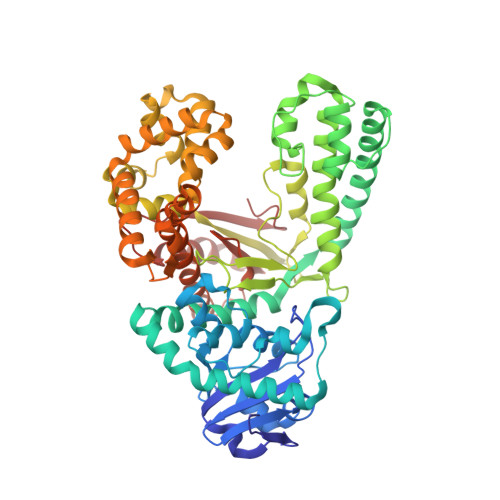
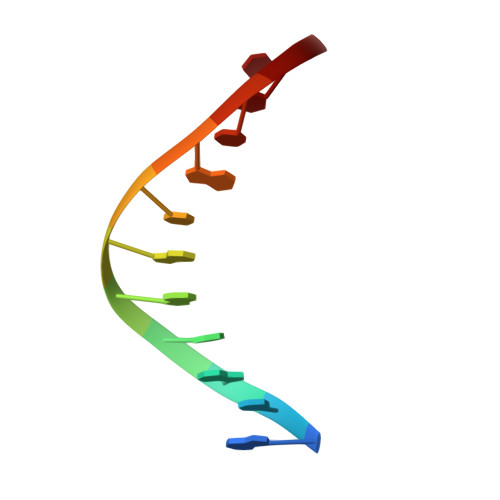
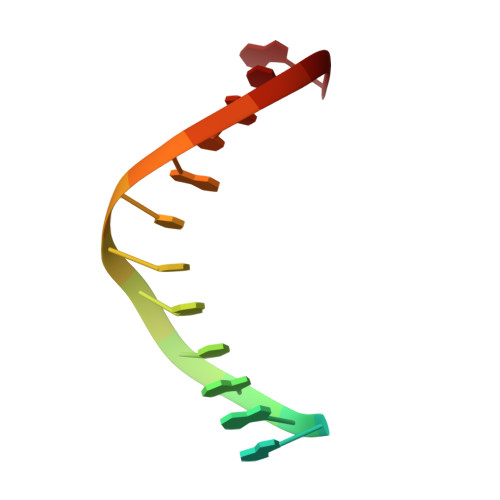
Structures of mismatch replication errors observed in a DNA polymerase.
Johnson, S.J., Beese, L.S.(2004) Cell 116: 803-816
- PubMed: 15035983
- DOI: https://doi.org/10.1016/s0092-8674(04)00252-1
- Primary Citation of Related Structures:
1NJW, 1NJX, 1NJY, 1NJZ, 1NK0, 1NK4, 1NK5, 1NK6, 1NK7, 1NK8, 1NK9, 1NKB, 1NKC, 1NKE - PubMed Abstract:
Accurate DNA replication is essential for genomic stability. One mechanism by which high-fidelity DNA polymerases maintain replication accuracy involves stalling of the polymerase in response to covalent incorporation of mismatched base pairs, thereby favoring subsequent mismatch excision. Some polymerases retain a "short-term memory" of replication errors, responding to mismatches up to four base pairs in from the primer terminus. Here we a present a structural characterization of all 12 possible mismatches captured at the growing primer terminus in the active site of a polymerase. Our observations suggest four mechanisms that lead to mismatch-induced stalling of the polymerase. Furthermore, we have observed the effects of extending a mismatch up to six base pairs from the primer terminus and find that long-range distortions in the DNA transmit the presence of the mismatch back to the enzyme active site, suggesting the structural basis for the short-term memory of replication errors.
Organizational Affiliation:
Department of Biochemistry, Duke University Medical Center, Durham, NC 27710, USA.