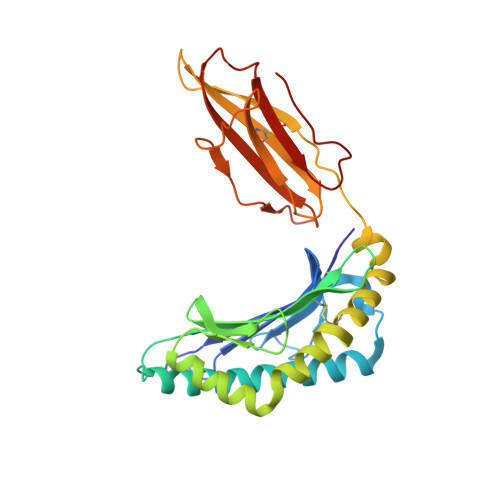
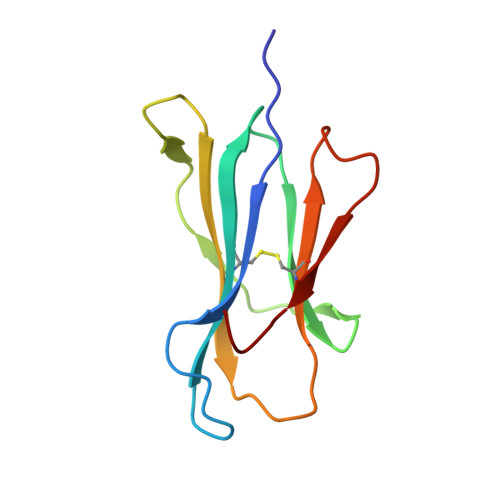
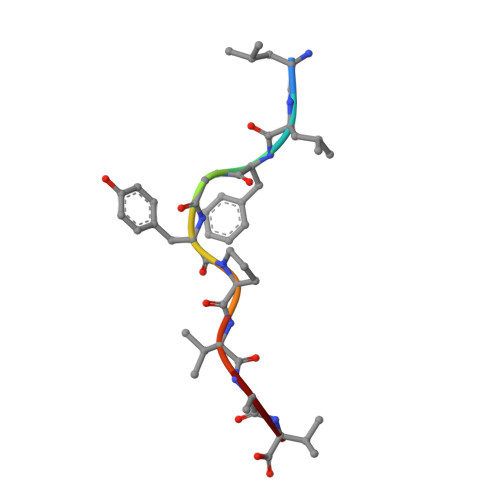
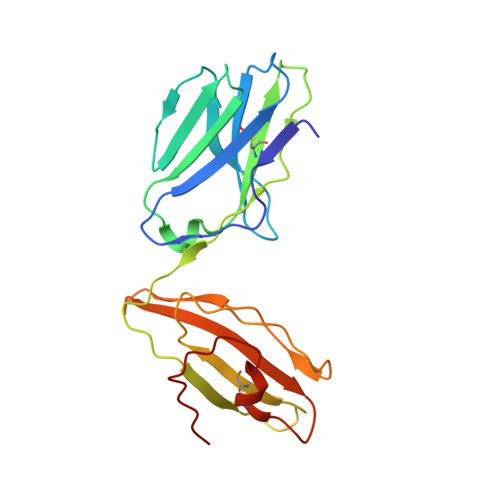
Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical.
Ding, Y.H., Baker, B.M., Garboczi, D.N., Biddison, W.E., Wiley, D.C.(1999) Immunity 11: 45-56
- PubMed: 10435578
- DOI: https://doi.org/10.1016/s1074-7613(00)80080-1
- Primary Citation of Related Structures:
1QRN, 1QSE, 1QSF - PubMed Abstract:
The interactions of three singly substituted peptide variants of the HTLV-1 Tax peptide bound to HLA-A2 with the A6 T cell receptor have been studied using T cell assays, kinetic and thermodynamic measurements, and X-ray crystallography. The three peptide/MHC ligands include weak agonists and antagonists with different affinities for TCR. The three-dimensional structures of the three A6-TCR/peptide/HLA-A2 complexes are remarkably similar to each other and to the wild-type agonist complex, with minor adjustments at the interface to accommodate the peptide substitutions (P6A, V7R, and Y8A). The lack of correlation between structural changes and the type of T cell signals induced provides direct evidence that different signals are not generated by different ligand-induced conformational changes in the alphabeta TCR.
Organizational Affiliation:
Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts 02138, USA.