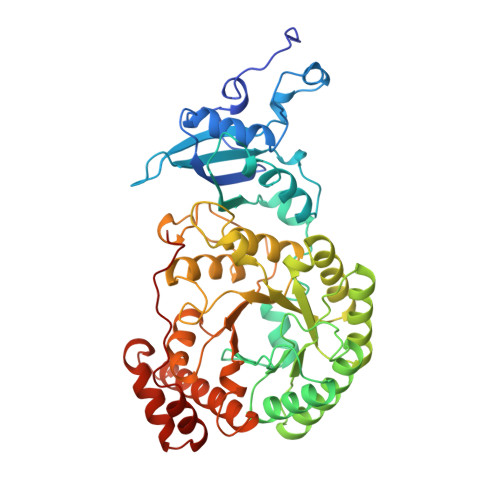
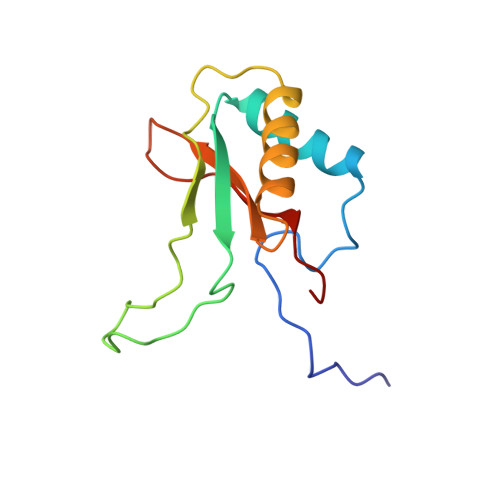
A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate.
Taylor, T.C., Fothergill, M.D., Andersson, I.(1996) J Biol Chem 271: 32894-32899
- PubMed: 8955130
- DOI: https://doi.org/10.1074/jbc.271.51.32894
- Primary Citation of Related Structures:
1RBO, 1RCO - PubMed Abstract:
Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) catalyzes the carboxylation of ribulose 1,5-bisphosphate. The reaction catalyzed by Rubisco involves several steps, some of which can occur as partial reactions, forming intermediates that can be isolated. Analogues of these intermediates are potent inhibitors of the enzyme. We have studied the interactions with the enzyme of two inhibitors, xylulose 1,5-bisphosphate and 4-carboxyarabinitol 1,5-bisphosphate, by x-ray crystallography. Crystals of the complexes were formed by cocrystallization under activating conditions. In addition, 4-carboxyarabinitol 1,5-bisphosphate was soaked into preformed activated crystals of the enzyme. The result of these experiments was the release of the activating CO2 molecule as well as the metal ion from the active site when the inhibitors bound to the enzyme. Comparison with the structure of an activated complex of the enzyme indicates that the structural basis for the release of the activator groups is a distortion of the metal binding site due to the different geometry of the C-3 hydroxyl of the inhibitors. Both inhibitors induce closure of active site loops despite the inactivated state of the enzyme. Xylulose 1,5-bisphosphate binds in a hydrated form at the active site.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, P. O. Box 590, S-751 24 Uppsala, Sweden.