Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
Webb, A.I., Dunstone, M.A., Chen, W., Aguilar, M.I., Chen, Q., Jackson, H., Chang, L., Kjer-Nielsen, L., Beddoe, T., McCluskey, J., Rossjohn, J., Purcell, A.W.(2004) J Biol Chem 279: 23438-23446
- PubMed: 15004033
- DOI: https://doi.org/10.1074/jbc.M314066200
- Primary Citation of Related Structures:
1S9W, 1S9X, 1S9Y - PubMed Abstract:
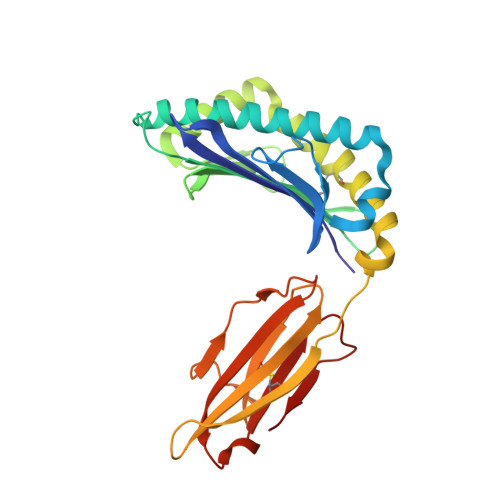
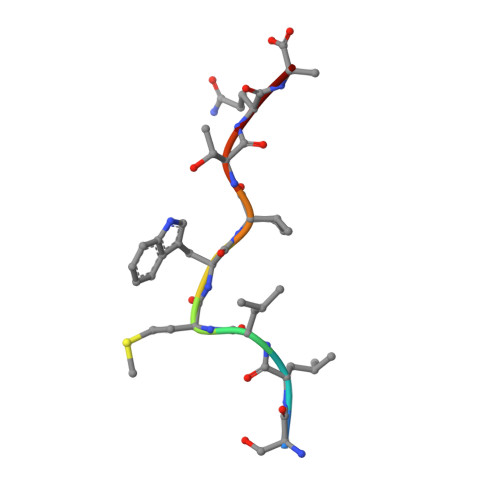
NY-ESO-1, a commonly expressed tumor antigen of the cancer-testis family, is expressed by a wide range of tumors but not found in normal adult somatic tissue, making it an ideal cancer vaccine candidate. Peptides derived from NY-ESO-1 have shown preclinical and clinical trial promise; however, biochemical features of these peptides have complicated their formulation and led to heterogeneous immune responses. We have taken a rational approach to engineer an HLA A2-restricted NY-ESO-1-derived T cell epitope with improved formulation and immunogenicity to the wild type peptide. To accomplish this, we have solved the x-ray crystallographic structures of HLA A2 complexed to NY-ESO (157-165) and two analogues of this peptide in which the C-terminal cysteine residue has been substituted to alanine or serine. Substitution of cysteine by serine maintained peptide conformation yet reduced complex stability, resulting in poor cytotoxic T lymphocyte recognition. Conversely, substitution with alanine maintained complex stability and cytotoxic T lymphocyte recognition. Based on the structures of the three HLA A2 complexes, we incorporated 2-aminoisobutyric acid, an isostereomer of cysteine, into the epitope. This analogue is impervious to oxidative damage, cysteinylation, and dimerization of the peptide epitope upon formulation that is characteristic of the wild type peptide. Therefore, this approach has yielded a potential therapeutic molecule that satiates the hydrophobic F pocket of HLA A2 and exhibited superior immunogenicity relative to the wild type peptide.
Organizational Affiliation:
Protein Crystallography Unit and Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.