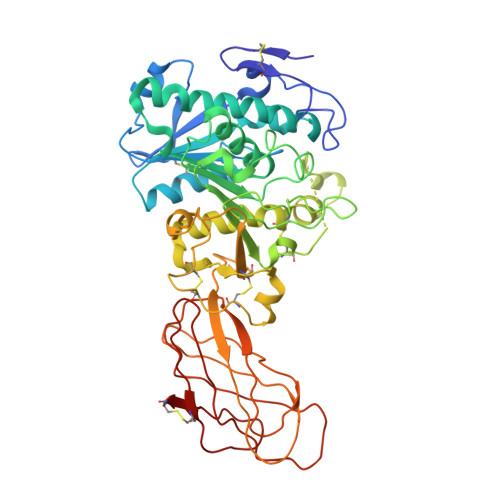
Crystalization of a Proteolyzed Form of the Horse Pancreatic Lipase-Related Protein 2: Structural Basis for the Specific Detergent Requirement.
Mancheno, J.M., Jayne, S., Kerfelec, B., Chapus, C., Crenon, I., Hermoso, J.A.(2004) Acta Crystallogr D Biol Crystallogr 60: 2107
- PubMed: 15502342
- DOI: https://doi.org/10.1107/S0907444904024229
- Primary Citation of Related Structures:
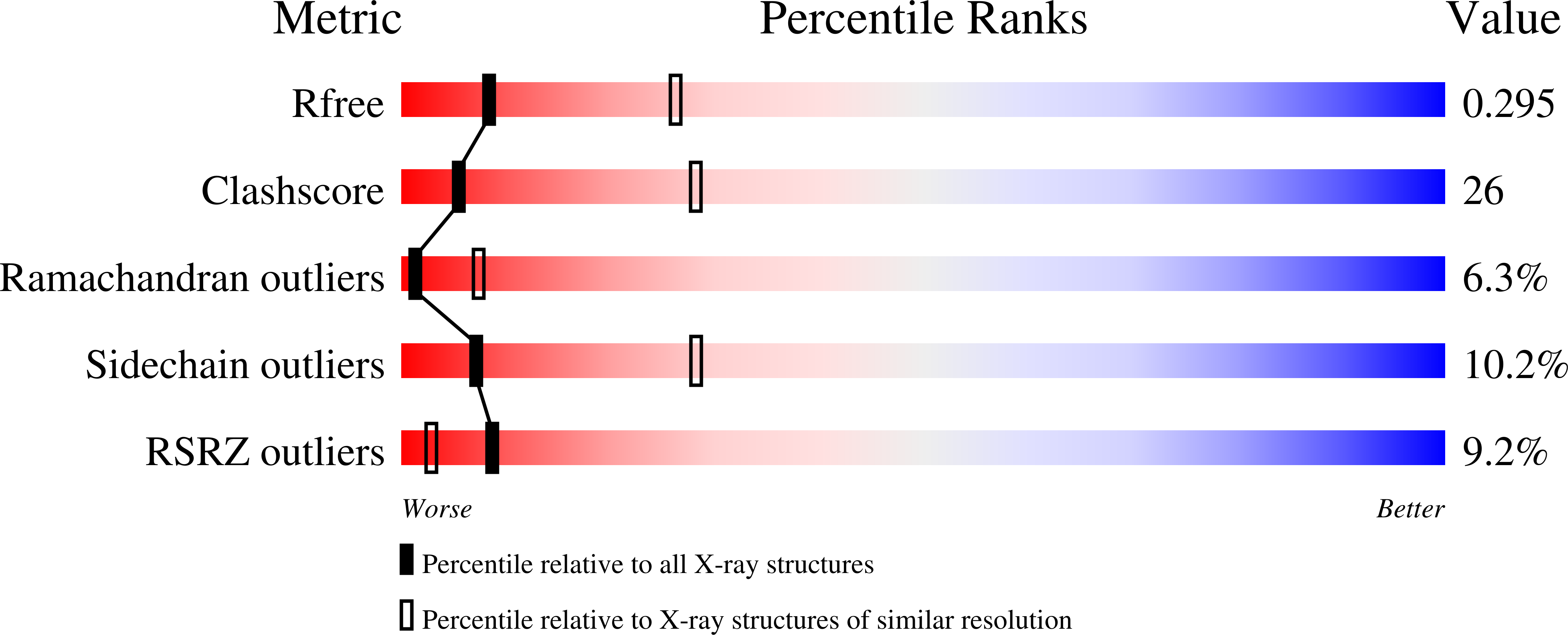
1W52 - PubMed Abstract:
Horse pancreatic lipase-related proteins PLRP1 and PLRP2 are produced by the pancreas together with pancreatic lipase (PL). Sequence-comparison analyses reveal that the three proteins possess the same two-domain organization: an N-terminal catalytic domain and a C-terminal domain, which in PL is involved in colipase binding. Nevertheless, despite the high level of sequence identity found, they exhibit distinct enzymatic properties. The intrinsic sensitivity of the peptide bond between Ser245 and Thr246 within the flap region of PLRP2 to proteolytic cleavage probably complicates PLRP2 crystallization since, as shown here, this proteolyzed form of PLRP2 is only crystallized after specific detergent stabilization of this region. This has been performed by the hanging-drop vapour-diffusion method at 291 K and exclusively in the presence of N,N-dimethyldecylamine-beta-oxide (DDAO). However, most crystals (>95%) are highly twinned and diffract poorly (to approximately 7-5 A resolution). Diffraction-quality trigonal crystals have unit-cell parameters a = b = 128.4, c = 85.8 A and belong to space group P3(2)21. A 2.9 A native data set was collected at ESRF on beamline ID14-2 with an R(merge) of 12.7%. Preliminary structural analysis provides a structural basis for the specific roles of DDAO.
Organizational Affiliation:
Grupo de Cristalografía Macromolecular y Biología Estructural, Instituto Rocasolano, CSIC, Serrano 119, 28006 Madrid, Spain. xjosemi@iqfr.csic.es