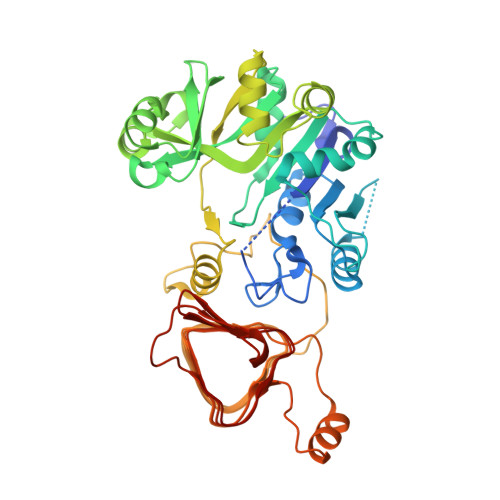
Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Jin, X., Ballicora, M.A., Preiss, J., Geiger, J.H.(2005) EMBO J 24: 694-704
- PubMed: 15692569
- DOI: https://doi.org/10.1038/sj.emboj.7600551
- Primary Citation of Related Structures:
1YP2, 1YP3, 1YP4 - PubMed Abstract:
ADP-glucose pyrophosphorylase catalyzes the first committed and rate-limiting step in starch biosynthesis in plants and glycogen biosynthesis in bacteria. It is the enzymatic site for regulation of storage polysaccharide accumulation in plants and bacteria, being allosterically activated or inhibited by metabolites of energy flux. We report the first atomic resolution structure of ADP-glucose pyrophosphorylase. Crystals of potato tuber ADP-glucose pyrophosphorylase alpha subunit were grown in high concentrations of sulfate, resulting in the sulfate-bound, allosterically inhibited form of the enzyme. The N-terminal catalytic domain resembles a dinucleotide-binding Rossmann fold and the C-terminal domain adopts a left-handed parallel beta helix that is involved in cooperative allosteric regulation and a unique oligomerization. We also report structures of the enzyme in complex with ATP and ADP-glucose. Communication between the regulator-binding sites and the active site is both subtle and complex and involves several distinct regions of the enzyme including the N-terminus, the glucose-1-phosphate-binding site, and the ATP-binding site. These structures provide insights into the mechanism for catalysis and allosteric regulation of the enzyme.
Organizational Affiliation:
Department of Chemistry, Michigan State University, East Lansing, MI 48824, USA.