Structural analysis of ligand binding and catalysis in chorismate lyase
Smith, N.N., Roitberg, A.E., Rivera, E., Howard, A., Holden, M.J., Mayhew, M., Kaistha, S., Gallagher, D.T.(2006) Arch Biochem Biophys 445: 72-80
- PubMed: 16343413
- DOI: https://doi.org/10.1016/j.abb.2005.10.026
- Primary Citation of Related Structures:
1JD3, 1TT8, 1XLR, 2AHC - PubMed Abstract:
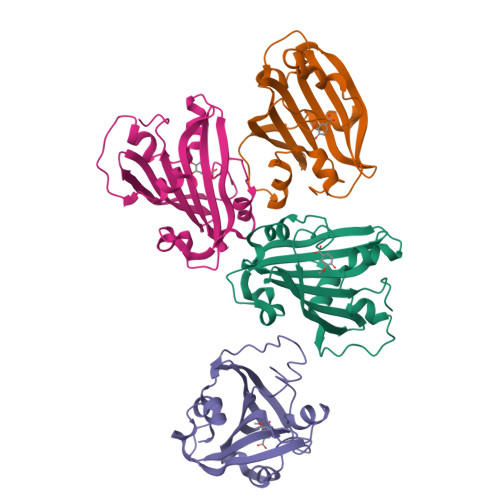
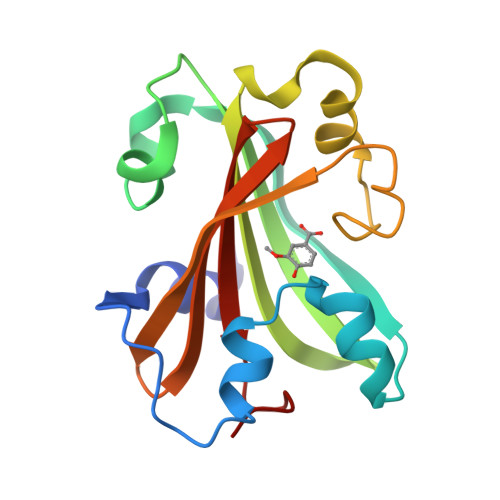
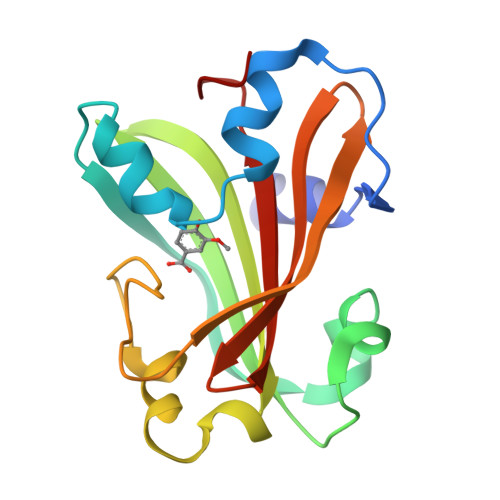
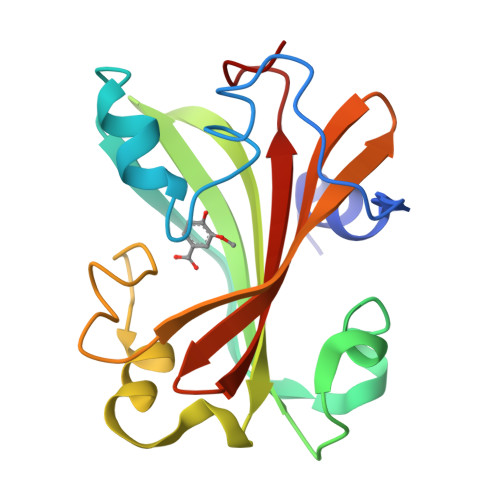
Chorismate lyase (CL) removes the pyruvyl group from chorismate to provide 4-hydroxybenzoate (4HB) for the ubiquinone pathway. We previously reported the crystal structure at 1.4A resolution of the Escherichia coli CL with bound 4HB product, showing that the product is bound in an internal cavity behind two flaps. To provide a more complete basis for understanding CL's unusual ligand-binding properties and mechanism of action, we now report four crystal structures of CL mutants and inhibitor complexes, together with binding and activity measurements and molecular dynamics simulations. First, an ultrahigh resolution (1.0A) crystal structure of the CL*product complex reveals details of a substrate-sized internal cavity, also behind the flaps, near the product site. Second, a 2.4A structure of CL complexed with the inhibitor vanillate shows the flaps partly opened relative to their product-bound positions. Third, a 2.0A structure of the G90A mutant with bound product reveals the basis for tighter product binding and kinetic effects of this active site mutation. Fourth, the combination of the G90A mutation with the vanillate inhibitor produces a 1.9A structure containing two inhibitor molecules, one in the product site and the other in the adjacent cavity. The two sites are connected by a short tunnel that is partly open at each end, suggesting that CL may operate via a 2-site or tunnel mechanism.
Organizational Affiliation:
University of Maryland Biotechnology Institute, Rockville, MD 20850, USA.