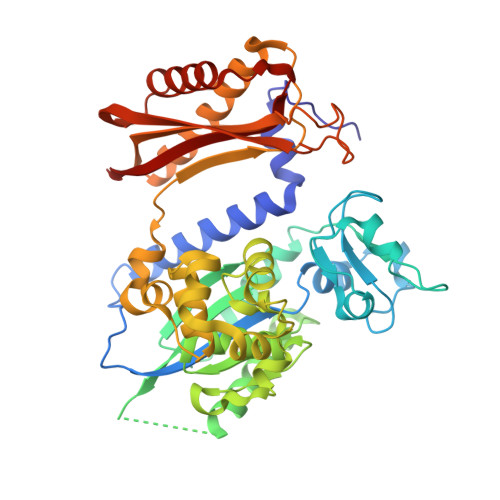
Rev1 employs a novel mechanism of DNA synthesis using a protein template.
Nair, D.T., Johnson, R.E., Prakash, L., Prakash, S., Aggarwal, A.K.(2005) Science 309: 2219-2222
- PubMed: 16195463
- DOI: https://doi.org/10.1126/science.1116336
- Primary Citation of Related Structures:
2AQ4 - PubMed Abstract:
The Rev1 DNA polymerase is highly specialized for the incorporation of C opposite template G. We present here the crystal structure of yeast Rev1 bound to template G and incoming 2'-deoxycytidine 5'-triphosphate (dCTP), which reveals that the polymerase itself dictates the identity of the incoming nucleotide, as well as the identity of the templating base. Template G and incoming dCTP do not pair with each other. Instead, the template G is evicted from the DNA helix, and it makes optimal hydrogen bonds with a segment of Rev1. Also, unlike other DNA polymerases, incoming dCTP pairs with an arginine rather than the templating base, which ensures the incorporation of dCTP over other incoming nucleotides. This mechanism provides an elegant means for promoting proficient and error-free synthesis through N2-adducted guanines that obstruct replication.
Organizational Affiliation:
Structural Biology Program, Department of Physiology and Biophysics, Mount Sinai School of Medicine, Box 1677, 1425 Madison Avenue, New York, NY 10029, USA.