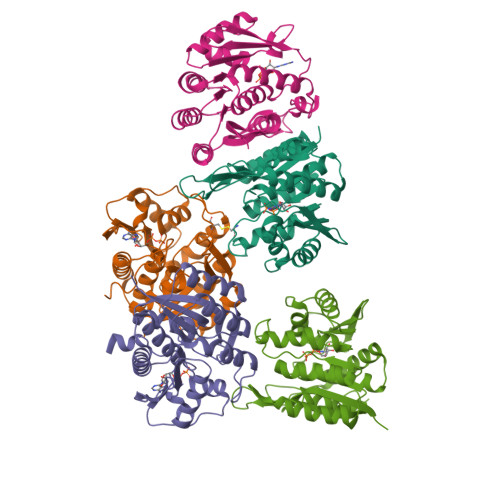
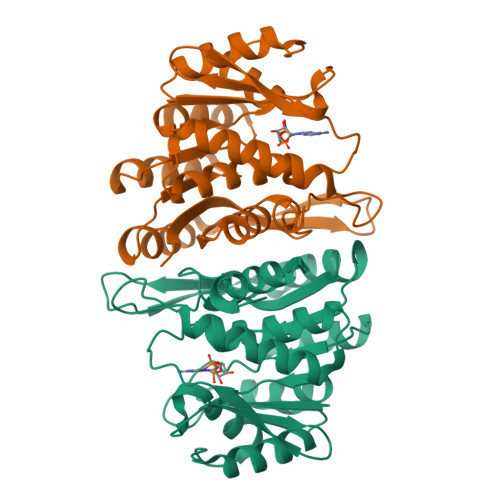
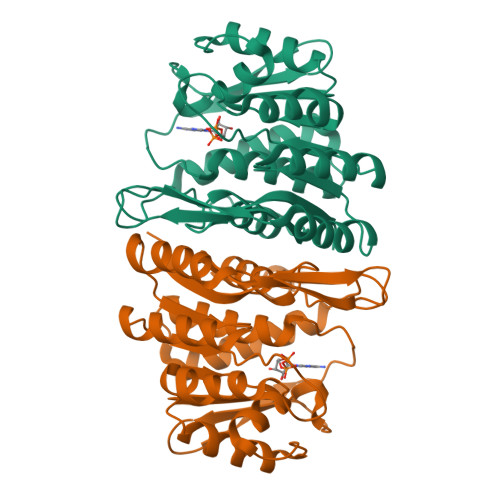
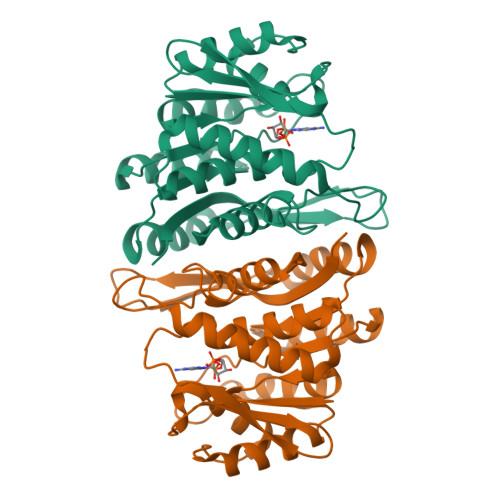
The Crystal Structure of an ADP Complex of Bacillus subtilis Pyridoxal Kinase Provides Evidence for the Parallel Emergence of Enzyme Activity During Evolution.
Newman, J.A., Das, S.K., Sedelnikova, S.E., Rice, D.W.(2006) J Mol Biology 363: 520-530
- PubMed: 16978644
- DOI: https://doi.org/10.1016/j.jmb.2006.08.013
- Primary Citation of Related Structures:
2I5B - PubMed Abstract:
Pyridoxal kinase catalyses the phosphorylation of pyridoxal, pyridoxine and pyridoxamine to their 5' phosphates and plays an important role in the pyridoxal 5' phosphate salvage pathway. The crystal structure of a dimeric pyridoxal kinase from Bacillus subtilis has been solved in complex with ADP to 2.8 A resolution. Analysis of the structure suggests that binding of the nucleotide induces the ordering of two loops, which operate independently to close a flap on the active site. Comparisons with other ribokinase superfamily members reveal that B. subtilis pyridoxal kinase is more closely related in both sequence and structure to the family of HMPP kinases than to other pyridoxal kinases, suggesting that this structure represents the first for a novel family of "HMPP kinase-like" pyridoxal kinases. Moreover this further suggests that this enzyme activity has evolved independently on multiple occasions from within the ribokinase superfamily.
Organizational Affiliation:
Krebs Institute for Biomolecular Research, Department of Molecular Biology and Biotechnology, The University of Sheffield, Sheffield S10 2TN, UK.