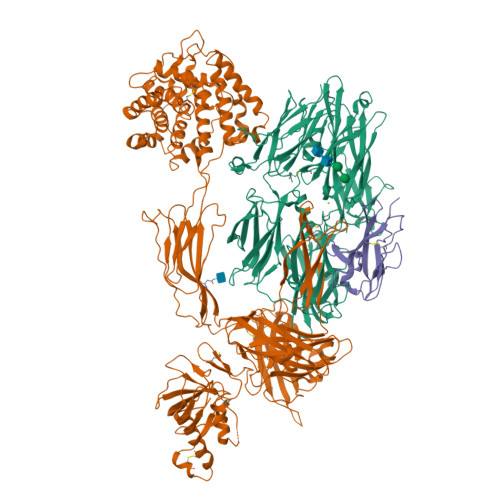
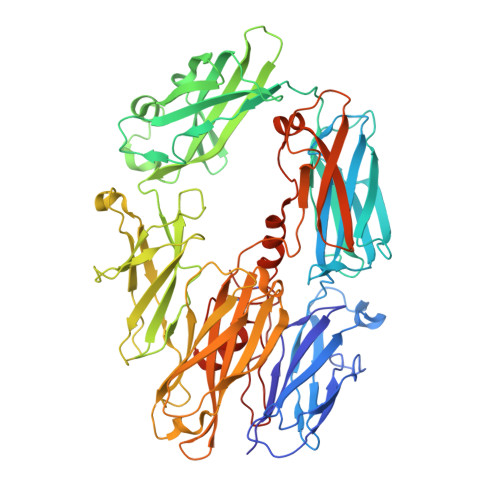
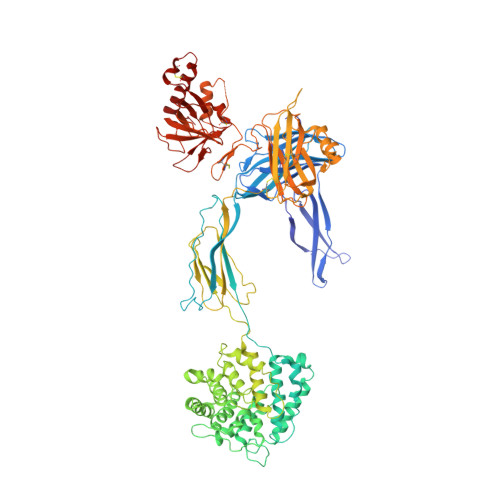
Structure of C3b in complex with CRIg gives insights into regulation of complement activation.
Wiesmann, C., Katschke, K.J., Yin, J., Helmy, K.Y., Steffek, M., Fairbrother, W.J., McCallum, S.A., Embuscado, L., DeForge, L., Hass, P.E., van Lookeren Campagne, M.(2006) Nature 444: 217-220
- PubMed: 17051150
- DOI: https://doi.org/10.1038/nature05263
- Primary Citation of Related Structures:
2ICC, 2ICE, 2ICF - PubMed Abstract:
The complement system is a key part of the innate immune system, and is required for clearance of pathogens from the bloodstream. After exposure to pathogens, the third component of the complement system, C3, is cleaved to C3b which, after recruitment of factor B, initiates formation of the alternative pathway convertases. CRIg, a complement receptor expressed on macrophages, binds to C3b and iC3b mediating phagocytosis of the particles, but it is unknown how CRIg selectively recognizes proteolytic C3-fragments and whether binding of CRIg to C3b inhibits convertase activation. Here we present the crystal structure of C3b in complex with CRIg and, using CRIg mutants, provide evidence that CRIg acts as an inhibitor of the alternative pathway of complement. The structure shows that activation of C3 induces major structural rearrangements, including a dramatic movement (>80 A) of the thioester-bond-containing domain through which C3b attaches to pathogen surfaces. We show that CRIg is not only a phagocytic receptor, but also a potent inhibitor of the alternative pathway convertases. The structure provides insights into the complex macromolecular structural rearrangements that occur during complement activation and inhibition. Moreover, our structure-function studies relating the structural basis of complement activation and the means by which CRIg inhibits the convertases provide important clues to the development of therapeutics that target complement.
Organizational Affiliation:
Department of Protein Engineering, 1 DNA Way, South San Francisco, California 94080, USA.