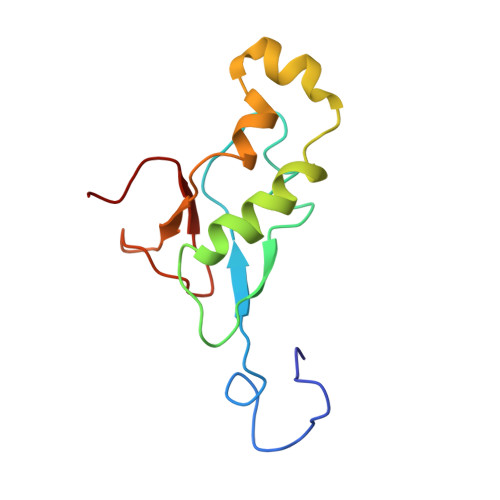
Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
Serrano, P., Johnson, M.A., Almeida, M.S., Horst, R., Herrmann, T., Joseph, J.S., Neuman, B.W., Subramanian, V., Saikatendu, K.S., Buchmeier, M.J., Stevens, R.C., Kuhn, P., Wuthrich, K.(2007) J Virol 81: 12049-12060
- PubMed: 17728234
- DOI: https://doi.org/10.1128/JVI.00969-07
- Primary Citation of Related Structures:
2GRI, 2IDY - PubMed Abstract:
This paper describes the structure determination of nsp3a, the N-terminal domain of the severe acute respiratory syndrome coronavirus (SARS-CoV) nonstructural protein 3. nsp3a exhibits a ubiquitin-like globular fold of residues 1 to 112 and a flexibly extended glutamic acid-rich domain of residues 113 to 183. In addition to the four beta-strands and two alpha-helices that are common to ubiquitin-like folds, the globular domain of nsp3a contains two short helices representing a feature that has not previously been observed in these proteins. Nuclear magnetic resonance chemical shift perturbations showed that these unique structural elements are involved in interactions with single-stranded RNA. Structural similarities with proteins involved in various cell-signaling pathways indicate possible roles of nsp3a in viral infection and persistence.
Organizational Affiliation:
Department of Molecular Biology, MB-44, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.