Structure and Reaction Geometry of Geranylgeranyl Diphosphate Synthase from Sinapis Alba.
Kloer, D.P., Welsch, R., Beyer, P., Schulz, G.E.(2006) Biochemistry 45: 15197
- PubMed: 17176041
- DOI: https://doi.org/10.1021/bi061572k
- Primary Citation of Related Structures:
2J1O, 2J1P - PubMed Abstract:
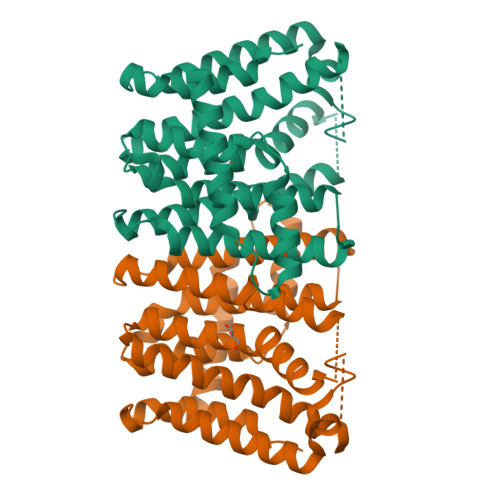
The crystal structure of the geranylgeranyl diphosphate synthase from Sinapis alba (mustard) has been solved in two crystal forms at 1.8 and 2.0 A resolutions. In one of these forms, the dimeric enzyme binds one molecule of the final product geranylgeranyl diphosphate in one subunit. The chainfold of the enzyme corresponds to that of other members of the farnesyl diphosphate synthase family. Whereas the binding modes of the two substrates dimethylallyl diphosphate and isopentenyl diphosphate at the allyl and isopentenyl sites, respectively, have been established with other members of the family, the complex structure presented reveals for the first time the binding mode of a reaction product at the isopentenyl site. The binding geometry of substrates and product in conjunction with the protein environment and the established chemistry of the reaction provide a clear picture of the reaction steps and atom displacements. Moreover, a comparison with a ligated homologous structure outlined an appreciable induced fit: helix alpha8 and its environment undergo a large conformational change when either the substrate dimethylallyl diphosphate or an analogue is bound to the allyl site; only a minor conformational change occurs when the other substrate isopentenyl diphosphate or the product is bound to the isopentenyl site.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie, Albert-Ludwigs-Universität, Albertstrasse 21, D-79104 Freiburg im Breisgau, Germany.