A New DNA Binding Protein Highly Conserved in Diverse Crenarchaeal Viruses
Larson, E.T., Eilers, B.J., Reiter, D., Ortmann, A.C., Young, M.J., Lawrence, C.M.(2007) Virology 363: 387
- PubMed: 17336360
- DOI: https://doi.org/10.1016/j.virol.2007.01.027
- Primary Citation of Related Structures:
2J85 - PubMed Abstract:
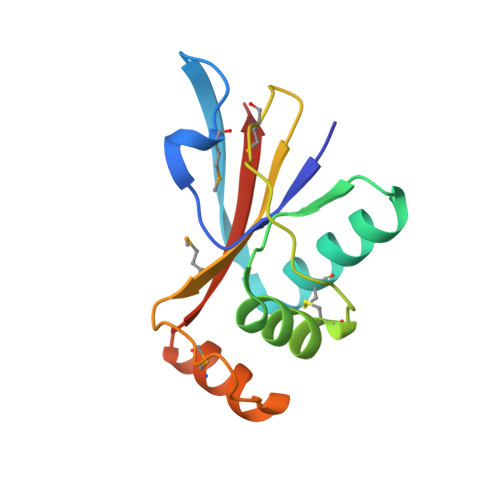
Sulfolobus turreted icosahedral virus (STIV) infects Sulfolobus species found in the hot springs of Yellowstone National Park. Its 37 open reading frames (ORFs) generally lack sequence similarity to other genes. One exception, however, is ORF B116. While its function is unknown, orthologs are found in three additional crenarchaeal viral families. Due to the central importance of this protein family to crenarchaeal viruses, we have undertaken structural and biochemical studies of B116. The structure reveals a previously unobserved fold consisting of a five-stranded beta-sheet flanked on one side by three alpha helices. Two subunits come together to form a homodimer with a 10-stranded mixed beta-sheet, where the topology of the central strands resembles an unclosed beta-barrel. Highly conserved loops rise above the surface of the saddle-shaped protein and suggest an interaction with the major groove of DNA. The predicted B116-DNA interaction is confirmed by electrophoretic mobility shift assays.
Organizational Affiliation:
Thermal Biology Institute, Montana State University, Bozeman, MT 59717, USA.