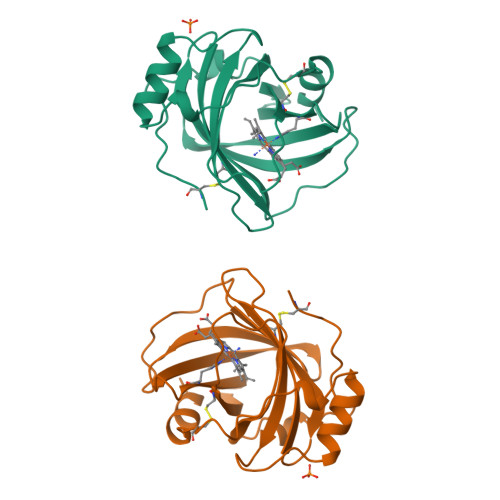
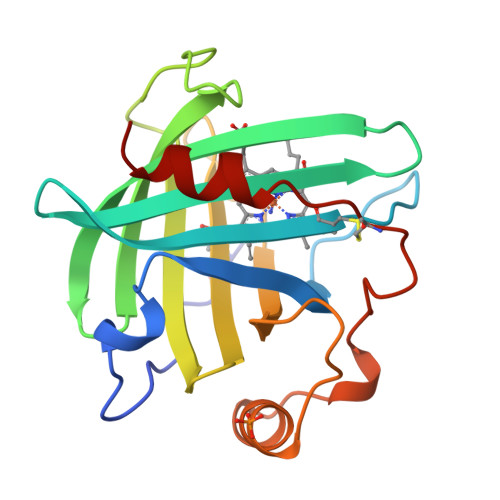
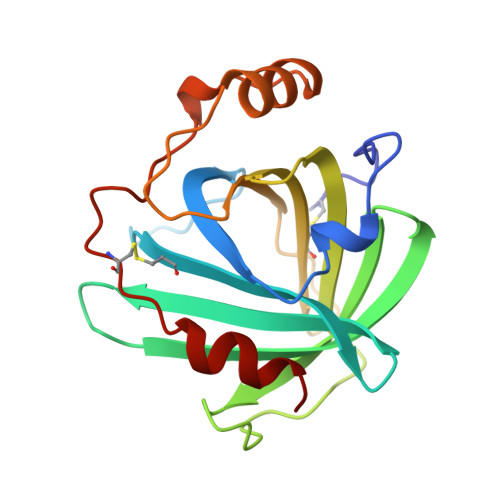
Crystal structures of a nitric oxide transport protein from a blood-sucking insect.
Weichsel, A., Andersen, J.F., Champagne, D.E., Walker, F.A., Montfort, W.R.(1998) Nat Struct Biol 5: 304-309
- PubMed: 9546222
- DOI: https://doi.org/10.1038/nsb0498-304
- Primary Citation of Related Structures:
1NP1, 2NP1, 3NP1 - PubMed Abstract:
The nitrophorins are heme-based proteins from the salivary glands of the blood-sucking insect Rhodnius prolixus that deliver nitric oxide gas (NO) to the victim while feeding, resulting in vasodilation and inhibition of platelet aggregation. The nitrophorins also bind tightly to histamine, which is released by the host to induce wound healing. Here we present three crystal structures of nitrophorin 1 (NP1): bound to cyanide, which binds in a manner similar to NO (2.3 A resolution); bound to histamine (2.0 A resolution); and bound to what appears to be NH3 from the crystallization solution (2.0 A resolution). The NP1 structures reveal heme to be sandwiched between strands of a lipocalin-like beta-barrel, and in an arrangement unlike any other gas-transport protein discovered to date. The heme is six-coordinate with a histidine (His 59) on the proximal side, and ligand in a spacious pocket on the distal side. The structures confirm that NO and histamine compete for the same binding pocket and become buried on binding. The dissociation constant for histamine binding was found to be 19 nM, approximately 100-fold lower than that for NO.
Organizational Affiliation:
Department of Biochemistry, University of Arizona, Tucson 85721, USA.