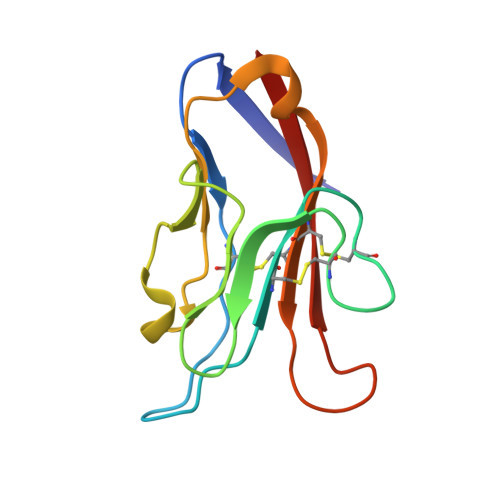
T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Cao, E., Zang, X., Ramagopal, U.A., Mukhopadhaya, A., Fedorov, A.A., Fedorov, E.V., Zencheck, W.D., Lary, J.W., Cole, J.L., Deng, H., Xiao, H., Dilorenzo, T.P., Allison, J.P., Nathenson, S.G., Almo, S.C.(2007) Immunity 26: 311-321
- PubMed: 17363302
- DOI: https://doi.org/10.1016/j.immuni.2007.01.016
- Primary Citation of Related Structures:
2OYP - PubMed Abstract:
The T cell immunoglobulin mucin (Tim) family of receptors regulates effector CD4(+) T cell functions and is implicated in autoimmune and allergic diseases. Tim-3 induces immunological tolerance, and engagement of the Tim-3 immunoglobulin variable (IgV) domain by galectin-9 is important for appropriate termination of T helper 1-immune responses. The 2 A crystal structure of the Tim-3 IgV domain demonstrated that four cysteines, which are invariant within the Tim family, form two noncanonical disulfide bonds, resulting in a surface not present in other immunoglobulin superfamily members. Biochemical and biophysical studies demonstrated that this unique structural feature mediates a previously unidentified galectin-9-independent binding process and suggested that this structural feature is conserved within the entire Tim family. The current work provided a graphic example of the relationship between sequence, structure, and function and suggested that the interplay between multiple Tim-3-binding activities contributes to the regulated assembly of signaling complexes required for effective Th1-mediated immunity.
Organizational Affiliation:
Department of Cell Biology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.