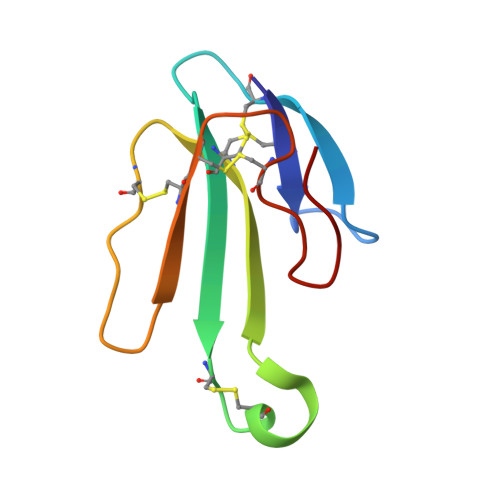
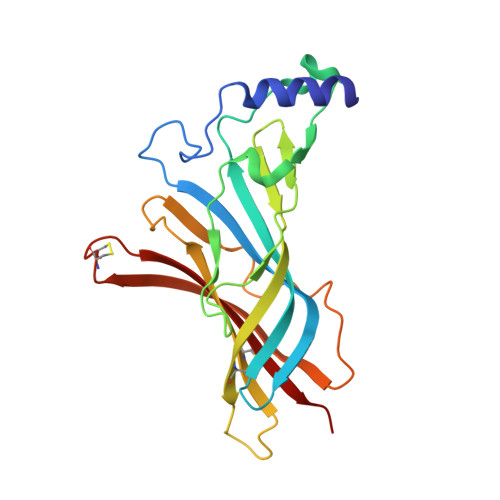
Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution.
Dellisanti, C.D., Yao, Y., Stroud, J.C., Wang, Z.Z., Chen, L.(2007) Nat Neurosci 10: 953-962
- PubMed: 17643119
- DOI: https://doi.org/10.1038/nn1942
- Primary Citation of Related Structures:
2QC1 - PubMed Abstract:
We determined the crystal structure of the extracellular domain of the mouse nicotinic acetylcholine receptor (nAChR) alpha1 subunit bound to alpha-bungarotoxin at 1.94 A resolution. This structure is the first atomic-resolution view of a nAChR subunit extracellular domain, revealing receptor-specific features such as the main immunogenic region (MIR), the signature Cys-loop and the N-linked carbohydrate chain. The toxin binds to the receptor through extensive protein-protein and protein-sugar interactions. To our surprise, the structure showed a well-ordered water molecule and two hydrophilic residues deep in the core of the alpha1 subunit. The two hydrophilic core residues are highly conserved in nAChRs, but correspond to hydrophobic residues in the nonchannel homolog acetylcholine-binding proteins. We carried out site-directed mutagenesis and electrophysiology analyses to assess the functional role of the glycosylation and the hydrophilic core residues. Our structural and functional studies show essential features of the nAChR and provide new insights into the gating mechanism.
Organizational Affiliation:
Molecular and Computation Biology, University of Southern California, 1050 Childs Way, RIH201, Los Angeles, California 90089-2910, USA.