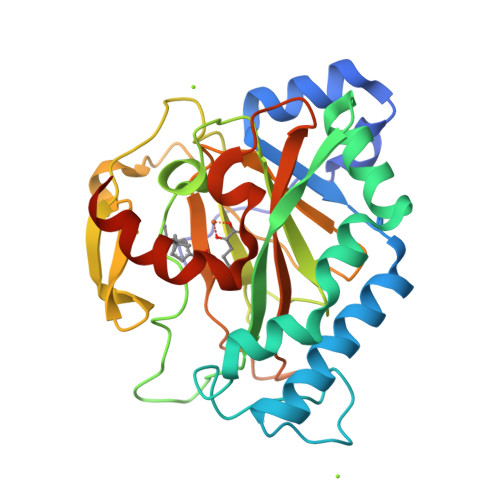
Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
You, Z., Omura, S., Ikeda, H., Cane, D.E., Jogl, G.(2007) J Biological Chem 282: 36552-36560
- PubMed: 17942405
- DOI: https://doi.org/10.1074/jbc.M706358200
- Primary Citation of Related Structures:
2RDN, 2RDQ, 2RDR, 2RDS - PubMed Abstract:
The non-heme iron dioxygenase PtlH from the soil organism Streptomyces avermitilis is a member of the iron(II)/alpha-ketoglutarate-dependent dioxygenase superfamily and catalyzes an essential reaction in the biosynthesis of the sesquiterpenoid antibiotic pentalenolactone. To investigate the structural basis for substrate recognition and catalysis, we have determined the x-ray crystal structure of PtlH in several complexes with the cofactors iron, alpha-ketoglutarate, and the non-reactive enantiomer of the substrate, ent-1-deoxypentalenic acid, in four different crystal forms to up to 1.31 A resolution. The overall structure of PtlH forms a double-stranded barrel helix fold, and the cofactor-binding site for iron and alpha-ketoglutarate is similar to other double-stranded barrel helix fold enzymes. Additional secondary structure elements that contribute to the substrate-binding site in PtlH are not conserved in other double-stranded barrel helix fold enzymes. Binding of the substrate enantiomer induces a reorganization of the monoclinic crystal lattice leading to a disorder-order transition of a C-terminal alpha-helix. The newly formed helix blocks the major access to the active site and effectively traps the bound substrate. Kinetic analysis of wild type and site-directed mutant proteins confirms a critical function of two arginine residues in substrate binding, while simulated docking of the enzymatic reaction product reveals the likely orientation of bound substrate.
Organizational Affiliation:
Department of Chemistry, Brown University, Providence, Rhode Island 02912-9108, USA.