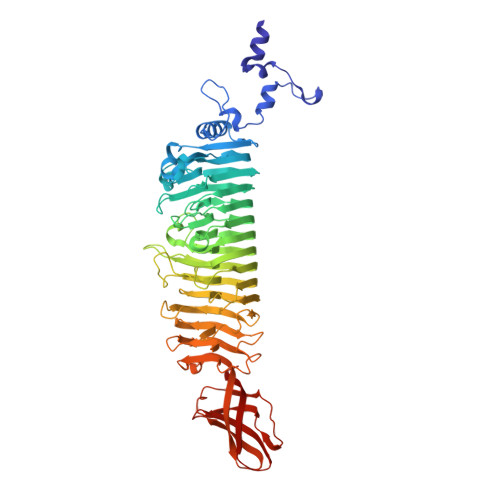
Crystal Structure of Escherichia Coli Phage Hk620 Tailspike: Podoviral Tailspike Endoglycosidase Modules are Evolutionarily Related.
Barbirz, S., Mueller, J.J., Uetrecht, C., Clark, A.J., Heinemann, U., Seckler, R.(2008) Mol Microbiol 69: 303
- PubMed: 18547389
- DOI: https://doi.org/10.1111/j.1365-2958.2008.06311.x
- Primary Citation of Related Structures:
2VJI, 2VJJ - PubMed Abstract:
Bacteriophage HK620 infects Escherichia coli H and is closely related to Shigella phage Sf6 and Salmonella phage P22. All three Podoviridae recognize and cleave their respective host cell receptor polysaccharide by homotrimeric tailspike proteins. The three proteins exhibit high sequence identity in the 110 residues of their N-terminal particle-binding domains, but no apparent sequence similarity in their major, receptor-binding parts. We have biochemically characterized the receptor-binding part of HK620 tailspike and determined its crystal structure to 1.38 A resolution. Its major domain is a right-handed parallel beta-helix, as in Sf6 and P22 tailspikes. HK620 tailspike has endo-N-acetylglucosaminidase activity and produces hexasaccharides of an O18A1-type O-antigen. As indicated by the structure of a hexasaccharide complex determined at 1.6 A resolution, the endoglycosidase-active sites are located intramolecularly, as in P22, and not between subunits, as in Sf6 tailspike. In contrast, the extreme C-terminal domain of HK620 tailspike forms a beta-sandwich, as in Sf6 and unlike P22 tailspike. Despite the different folds, structure-based sequence alignments of the C-termini reveal motifs conserved between the three proteins. We propose that the tailspike genes of P22, Sf6 and HK620 have a common precursor and are not mosaics of unrelated gene fragments.
Organizational Affiliation:
Physikalische Biochemie, Universität Potsdam, Karl-Liebknecht-Str. 24-25, 14476 Golm, Germany.