The Siderophore Binding Protein Feua Shows Limited Promiscuity Toward Exogenous Triscatecholates
Peuckert, F., Ramos-Vega, A.L., Miethke, M., Schwoerer, C.J., Albrecht, A.G., Oberthuer, M., Marahiel, M.A.(2011) Chem Biol 18: 907
- PubMed: 21802011
- DOI: https://doi.org/10.1016/j.chembiol.2011.05.006
- Primary Citation of Related Structures:
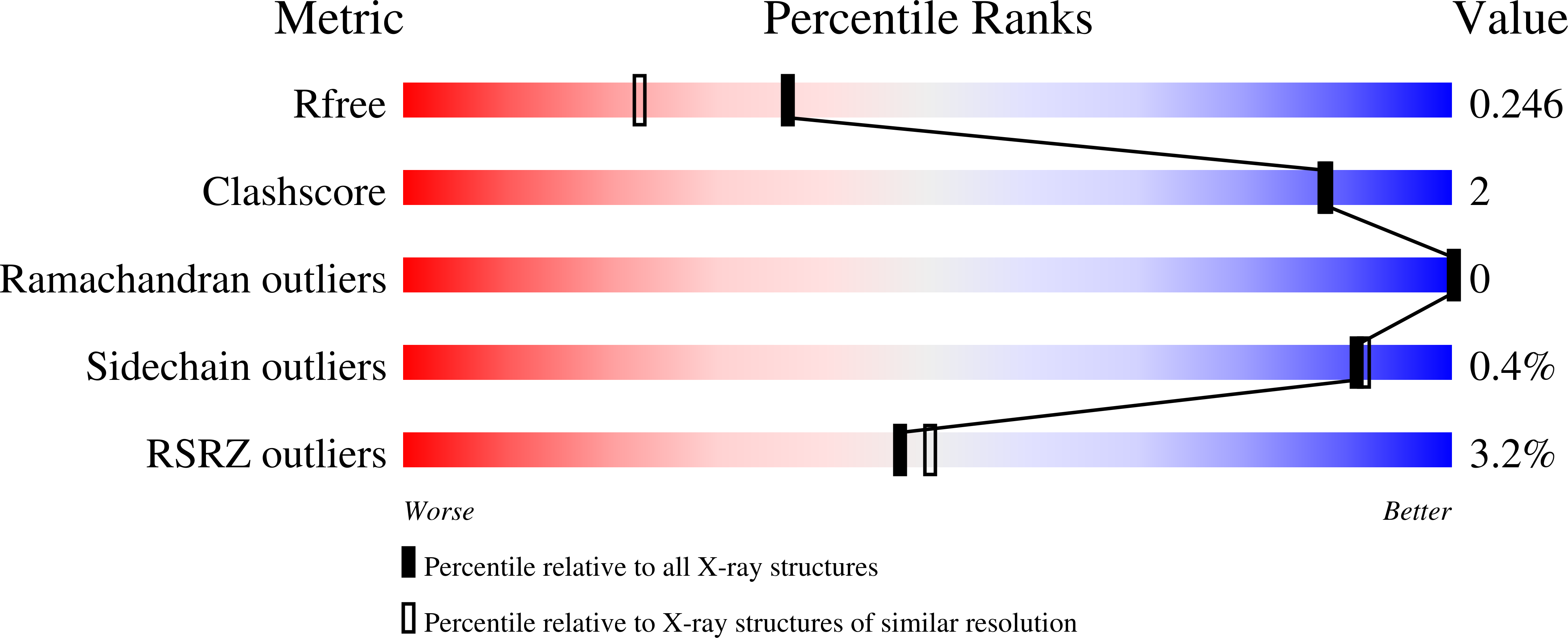
2XUZ, 2XV1 - PubMed Abstract:
Iron acquisition by siderophores is crucial for survival and virulence of many microorganisms. Here, we investigated the binding of the exogenous siderophore ferric enterobactin and the synthetic siderophore mimic ferric mecam by the triscatecholate binding protein FeuA from Bacillus subtilis at the atomic level. The structural complexes provide molecular insights into the capture mechanism of FeuA for exogenous and synthetic siderophores. The protein-ligand complexes show an exclusive acceptance of Λ-stereoconfigured substrates. Ligand-induced cross-bridging of the complexes was not observed, revealing a different thermodynamic behavior especially of the ferric mecam substrate, which was previously shown to dimerize with the enterobactin binding protein CeuE. The nearly identical overall domain movement of FeuA upon binding of ferric enterobactin or ferric mecam compared with endogenously derived ferric bacillibactin implies the importance of the conserved domain rearrangement for recognition by the transmembrane permease FeuBC, for which the conserved FeuA residues E90 and E221 were proved to be essential.
Organizational Affiliation:
Department of Chemistry, Biochemistry, Philipps-University Marburg, Hans-Meerwein-Straße, D-35032 Marburg, Germany.